Large Language Models Outperform Traditional Natural Language Processing Methods in Extracting Patient-Reported Outcomes in Inflammatory Bowel Disease
引用次数: 0
Abstract
Background and Aims
Patient-reported outcomes (PROs) are vital in assessing disease activity and treatment outcomes in inflammatory bowel disease (IBD). However, manual extraction of these PROs from the free-text of clinical notes is burdensome. We aimed to improve data curation from free-text information in the electronic health record, making it more available for research and quality improvement. This study aimed to compare traditional natural language processing (tNLP) and large language models (LLMs) in extracting 3 IBD PROs (abdominal pain, diarrhea, fecal blood) from clinical notes across 2 institutions.
Methods
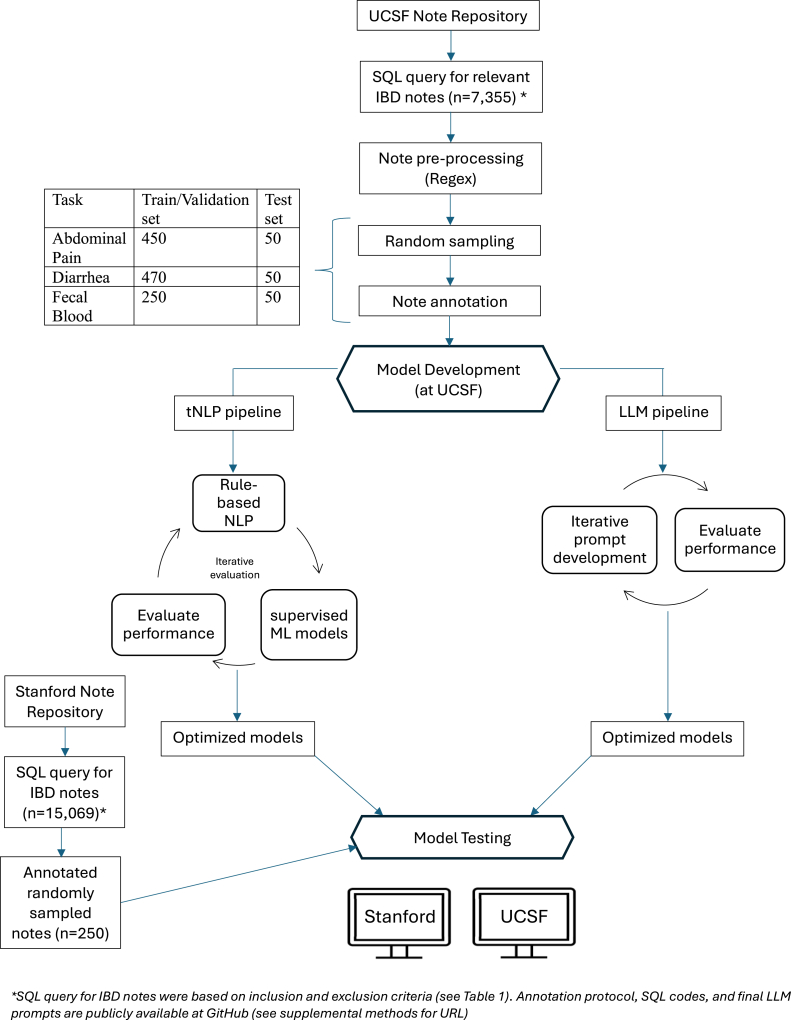
Clinic notes were annotated for each PRO using preset protocols. Models were developed and internally tested at the University of California, San Francisco, and then externally validated at Stanford University. We compared tNLP and LLM-based models on accuracy, sensitivity, specificity, positive, and negative predictive value. In addition, we conducted fairness and error assessments.
Results
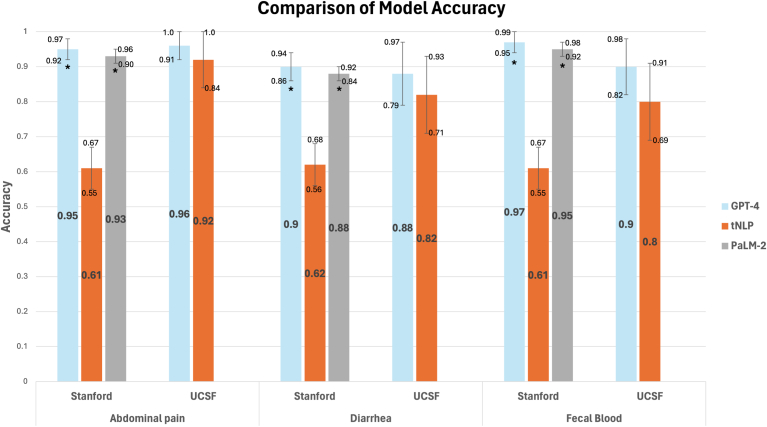
Interrater reliability between annotators was >90%. On the University of California, San Francisco test set (n = 50), the top-performing tNLP models showcased accuracies of 92% (abdominal pain), 82% (diarrhea) and 80% (fecal blood), comparable to GPT-4, which was 96%, 88%, and 90% accurate, respectively. On external validation at Stanford (n = 250), tNLP models failed to generalize (61%–62% accuracy) while GPT-4 maintained accuracies >90%. Pathways Language Model-2 and Generative Pre-trained Transformer-4 showed similar performance. No biases were detected based on demographics or diagnosis.
Conclusion
LLMs are accurate and generalizable methods for extracting PROs. They maintain excellent accuracy across institutions, despite heterogeneity in note templates and authors. Widespread adoption of such tools has the potential to enhance IBD research and patient care.
在提取炎症性肠病患者报告的结果方面,大型语言模型优于传统的自然语言处理方法。
背景和目的:患者报告的预后(PROs)对于评估炎症性肠病(IBD)的疾病活动性和治疗结果至关重要。然而,从临床记录的自由文本中手动提取这些优点是繁重的。我们的目标是改进电子健康记录中自由文本信息的数据管理,使其更易于研究和质量改进。本研究旨在比较传统自然语言处理(tNLP)和大型语言模型(LLMs)从两家机构的临床记录中提取3种IBD PROs(腹痛、腹泻、粪血)的效果。方法:采用预设方案对各PRO进行临床笔记注释。模型是在加州大学旧金山分校开发和内部测试的,然后在斯坦福大学进行外部验证。我们比较了tNLP和基于llm的模型的准确性、敏感性、特异性、阳性和阴性预测值。此外,我们还进行了公平性和误差评估。结果:注释者之间的互信度为90%。在加州大学旧金山分校的测试集(n = 50)上,表现最好的tNLP模型显示出92%(腹痛)、82%(腹泻)和80%(粪血)的准确率,与GPT-4相当,后者分别为96%、88%和90%。在斯坦福大学(n = 250)的外部验证中,tNLP模型未能泛化(准确率为61%-62%),而GPT-4保持了约90%的准确率。Pathways Language Model-2和Generative Pre-trained Transformer-4表现相似。没有发现基于人口统计学或诊断的偏差。结论:llm是一种准确、可推广的pro提取方法。尽管笔记模板和作者存在异质性,但它们在各机构之间保持了出色的准确性。这些工具的广泛采用有可能加强IBD研究和患者护理。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


