Synchronized long-read genome, methylome, epigenome and transcriptome profiling resolve a Mendelian condition
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
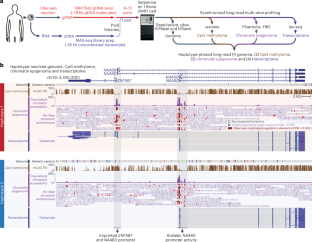
Resolving the molecular basis of a Mendelian condition remains challenging owing to the diverse mechanisms by which genetic variants cause disease. To address this, we developed a synchronized long-read genome, methylome, epigenome and transcriptome sequencing approach, which enables accurate single-nucleotide, insertion–deletion and structural variant calling and diploid de novo genome assembly. This permits the simultaneous elucidation of haplotype-resolved CpG methylation, chromatin accessibility and full-length transcript information in a single long-read sequencing run. Application of this approach to an Undiagnosed Diseases Network participant with a chromosome X;13-balanced translocation of uncertain significance revealed that this translocation disrupted the functioning of four separate genes (NBEA, PDK3, MAB21L1 and RB1) previously associated with single-gene Mendelian conditions. Notably, the function of each gene was disrupted via a distinct mechanism that required integration of the four ‘omes’ to resolve. These included fusion transcript formation, enhancer adoption, transcriptional readthrough silencing and inappropriate X-chromosome inactivation of autosomal genes. Overall, this highlights the utility of synchronized long-read multi-omic profiling for mechanistically resolving complex phenotypes. Simultaneous profiling of the genome, methylome, epigenome and transcriptome using single-molecule chromatin fiber sequencing and multiplexed arrays isoform sequencing identifies the genetic and molecular basis of an undiagnosed Mendelian disease case with an X;13-balanced translocation.

求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


