T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots
IF 27.7
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
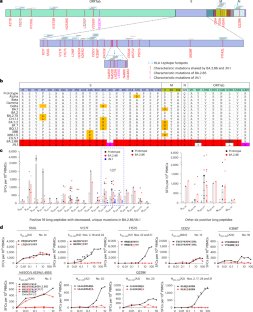
Although antibody escape is observed in emerging severe acute respiratory syndrome coronavirus 2 variants, T cell escape, especially after the global circulation of BA.2.86/JN.1, is unexplored. Here we demonstrate that T cell evasion exists in epitope hotspots spanning BA.2.86/JN.1 mutations. The newly emerging Q229K at this conserved nucleocapsid protein site impairs HLA-A2 epitope hotspot recognition. The association between HLA-A24 convalescents and T cell immune escape points to the spike (S) protein epitope S448–456NYNYLYRLF, with multiple mutations from Delta to JN.1, including L452Q, L452R, F456L, N450D and L452W, and N450D, L452W and L455S. A cliff drop of immune responses was observed for S448–456NYNYRYRLF (Delta/BA.5.2) and S448–456NYDYWYRSF (JN.1), but with immune preservation of S448–456NYDYWYRLF (BA.2.86). Structural analyses showed that hydrophobicity exposure determines the pronounced escape of L452R and L455S mutants, which was further confirmed by T cell receptor binding. This study highlights the characteristics and molecular mechanisms of the T cell immune escape for JN.1 and provides new insights into understanding the dominant circulation of variants, from the viewpoint of cytotoxic T cell evasion. Liu and colleagues examine how severe acute respiratory syndrome coronavirus 2 variants evade CD8+ T cell epitope recognition.

针对两个细胞毒性T细胞表位热点的SARS-CoV-2 JN.1逃逸者的T细胞免疫逃避
虽然在新出现的严重急性呼吸综合征冠状病毒2变异中观察到抗体逃逸,但T细胞逃逸,特别是在BA.2.86/JN全循环后。1、是未开发的。在这里,我们证明T细胞逃避存在于BA.2.86/JN表位热点中。1突变。在这个保守的核衣壳蛋白位点上新出现的Q229K损害了HLA-A2表位热点识别。HLA-A24康复期与T细胞免疫逃逸的关联指向spike (S)蛋白表位S448-456NYNYLYRLF,从Delta到JN.1存在多个突变,包括L452Q、L452R、F456L、N450D和L452W,以及N450D、L452W和L455S。S448-456NYNYRYRLF (Delta/BA.5.2)和S448-456NYDYWYRSF (j .1)免疫应答急剧下降,但S448-456NYDYWYRLF (BA.2.86)免疫应答保持不变。结构分析表明,疏水性暴露决定了L452R和L455S突变体的明显逃逸,T细胞受体结合进一步证实了这一点。本研究从细胞毒性T细胞逃逸的角度,揭示了JN.1的T细胞免疫逃逸的特征和分子机制,为理解变异的主导循环提供了新的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


