Hospital toilets and drainage systems as a reservoir for a long-term polyclonal outbreak of clinical infections with multidrug-resistant Klebsiella oxytoca species complex
IF 1.9
Q3 INFECTIOUS DISEASES
引用次数: 0
Abstract
Background
Nosocomial outbreaks with multidrug-resistant bacteria with a probable reservoir in hospital toilets and drainage systems have been increasingly reported.
Aim
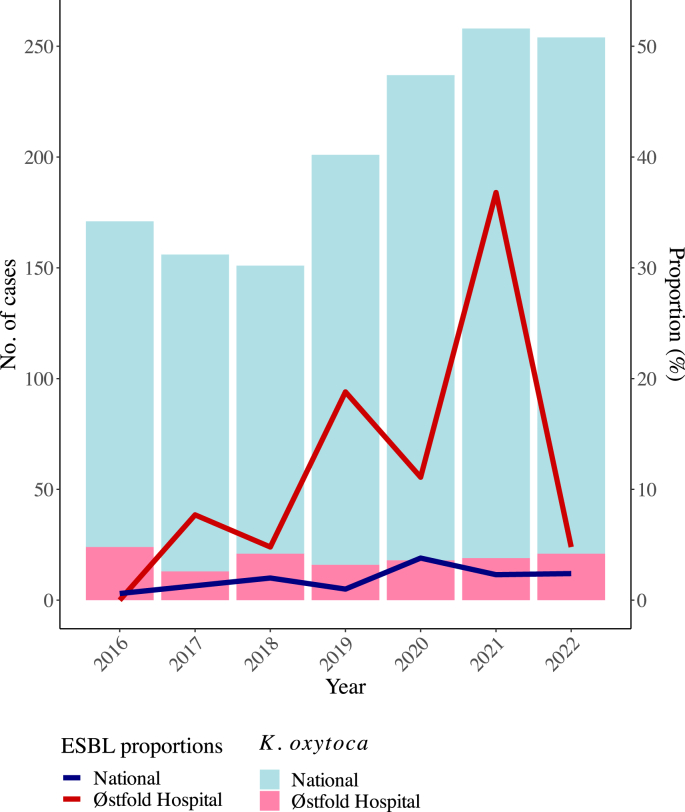
To investigate an increase in bacteraemia with extended-spectrum β-lactamase (ESBL)-producing Klebsiella oxytoca at our hospital in 2021; the epidemiology of the outbreak suggested an environmental source.
Methods
Available clinical K. oxytoca isolates from patient with infection or rectal carriage from 2019 to 2022 were collected. Clinical information was gathered from included patients and sampled sinks, shower drains, and toilet water. Short- and long-read whole-genome sequencing (WGS) was performed on patient and environmental isolates to assess phylogenetic relationships, antibiotic resistance genes/mutations, and plasmid profiles.
Results
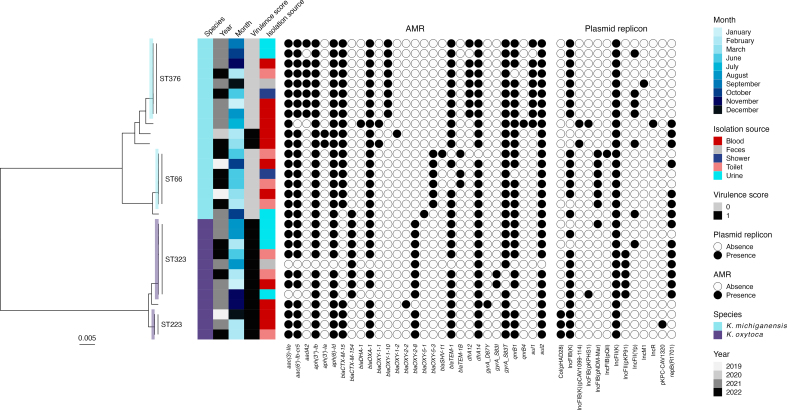
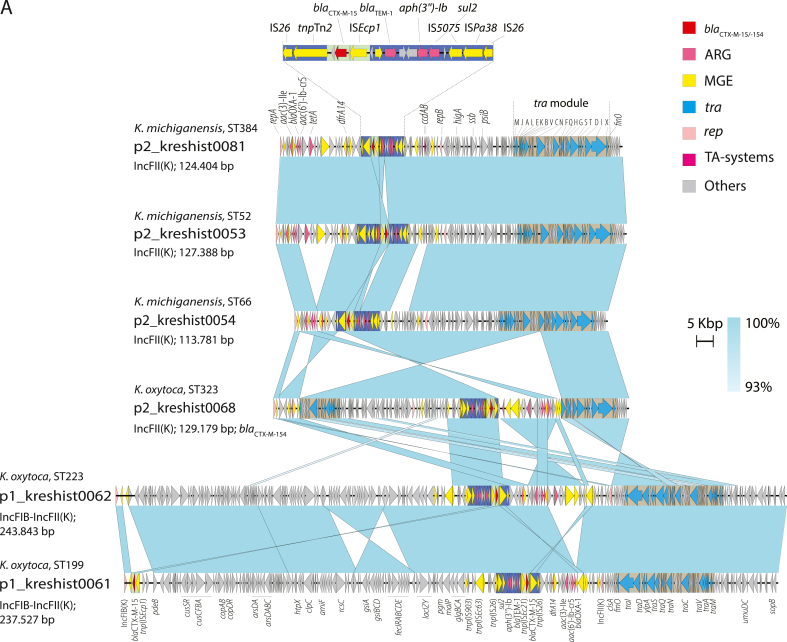
WGS revealed four clusters and a polyclonal population consisting of ESBL-producing K. oxytoca and Klebsiella michiganensis. All clusters contained both clinical and environmental isolates. The environmental sampling revealed widespread contamination of the outbreak strains in the outbreak ward, and plasmid analyses indicated possible transfer of plasmids between species and clones. Most environmental findings in the outbreak ward were from toilet water, and enhanced cleaning of bathrooms and toilets was introduced. The following year, a decrease in outbreak strains in systemic infections was observed.
Conclusion
This investigation uncovered a polyclonal outbreak of multidrug-resistant K. oxytoca and K. michiganensis and unveiled a persistent reservoir of outbreak clones in the drainage system and toilet water, facilitating exchange of resistance genes. The risk of toilet water as a source of clinical infections warrants further investigation.
医院厕所和排水系统是耐多药氧化克雷伯菌物种复合体临床感染长期多克隆爆发的蓄水池。
背景:医院内多药耐药菌暴发的报道越来越多,多药耐药菌可能存在于医院厕所和排水系统中。目的:了解我院2021年产β-内酰胺酶(ESBL)克雷伯菌菌血症的增加情况;疫情的流行病学分析表明这是环境因素造成的。方法:收集2019 - 2022年感染或直肠直肠携带患者临床分离的产氧克雷伯菌。从纳入的患者和取样的水槽、淋浴排水管和厕所用水中收集临床信息。对患者和环境分离株进行短读和长读全基因组测序(WGS),以评估系统发育关系、抗生素耐药基因/突变和质粒谱。结果:WGS检测到产esbl的oxytoca和Klebsiella michiganensis的4个聚类和多克隆群体。所有群集均含有临床和环境分离株。环境采样显示暴发病区暴发菌株受到广泛污染,质粒分析表明质粒可能在物种和克隆之间转移。疫情病房的大多数环境发现来自厕所水,并加强了浴室和厕所的清洁。次年,观察到全身感染的爆发菌株有所减少。结论:本次调查发现了多药耐药的oxytoca和k.m akanensis的多克隆爆发,并在排水系统和厕所水中发现了一个持续的爆发克隆库,促进了耐药基因的交换。厕所水作为临床感染源的风险值得进一步调查。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Infection Prevention in Practice
Medicine-Public Health, Environmental and Occupational Health
CiteScore
4.80
自引率
0.00%
发文量
58
审稿时长
61 days
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


