Spatially restricted and ontogenically distinct hepatic macrophages are required for tissue repair
IF 25.5
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
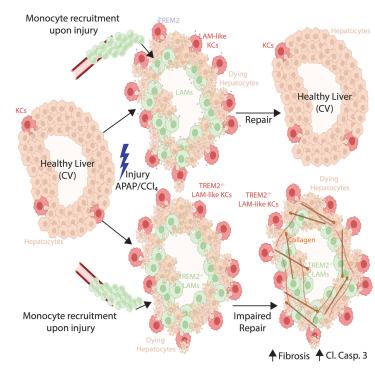
Our understanding of the functional heterogeneity of resident versus recruited macrophages in the diseased liver is limited. A population of recruited lipid-associated macrophages (LAMs) has been reported to populate the diseased liver alongside resident Kupffer cells (KCs). However, the precise roles of these distinct macrophage subsets remain elusive. Here, using proteogenomics, we have identified LAMs in multiple models of liver injury. Moreover, we found that this phenotype is not specific to recruited macrophages, as a subset of resident KCs can also adopt a LAM-like phenotype in the mouse and human liver. By combining genetic mouse models targeting the distinct populations, we determined that both recruited LAMs and resident LAM-like KCs play crucial roles in tissue repair. Specifically, triggering receptor expressed on myeloid cells 2 (TREM2) expression on either resident or recruited macrophages is required for the efficient clearance of dying cells, enhancing repair and preventing exacerbated fibrosis.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Immunity
医学-免疫学
CiteScore
49.40
自引率
2.20%
发文量
205
审稿时长
6 months
期刊介绍:
Immunity is a publication that focuses on publishing significant advancements in research related to immunology. We encourage the submission of studies that offer groundbreaking immunological discoveries, whether at the molecular, cellular, or whole organism level. Topics of interest encompass a wide range, such as cancer, infectious diseases, neuroimmunology, autoimmune diseases, allergies, mucosal immunity, metabolic diseases, and homeostasis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


