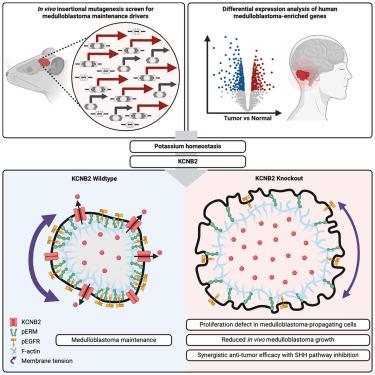
A forward genetic screen identifies potassium channel essentiality in SHH medulloblastoma maintenance
IF 10.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Distinguishing tumor maintenance genes from initiation, progression, and passenger genes is critical for developing effective therapies. We employed a functional genomic approach using the Lazy Piggy transposon to identify tumor maintenance genes in vivo and applied this to sonic hedgehog (SHH) medulloblastoma (MB). Combining Lazy Piggy screening in mice and transcriptomic profiling of human MB, we identified the voltage-gated potassium channel KCNB2 as a candidate maintenance driver. KCNB2 governs cell volume of MB-propagating cells (MPCs), with KCNB2 depletion causing osmotic swelling, decreased plasma membrane tension, and elevated endocytic internalization of epidermal growth factor receptor (EGFR), thereby mitigating proliferation of MPCs to ultimately impair MB growth. KCNB2 is largely dispensable for mouse development and KCNB2 knockout synergizes with anti-SHH therapy in treating MB. These results demonstrate the utility of the Lazy Piggy functional genomic approach in identifying cancer maintenance drivers and elucidate a mechanism by which potassium homeostasis integrates biomechanical and biochemical signaling to promote MB aggression.
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


