Ligand interaction landscape of transcription factors and essential enzymes in E. coli
IF 45.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Knowledge of protein-metabolite interactions can enhance mechanistic understanding and chemical probing of biochemical processes, but the discovery of endogenous ligands remains challenging. Here, we combined rapid affinity purification with precision mass spectrometry and high-resolution molecular docking to precisely map the physical associations of 296 chemically diverse small-molecule metabolite ligands with 69 distinct essential enzymes and 45 transcription factors in the gram-negative bacterium Escherichia coli. We then conducted systematic metabolic pathway integration, pan-microbial evolutionary projections, and independent in-depth biophysical characterization experiments to define the functional significance of ligand interfaces. This effort revealed principles governing functional crosstalk on a network level, divergent patterns of binding pocket conservation, and scaffolds for designing selective chemical probes. This structurally resolved ligand interactome mapping pipeline can be scaled to illuminate the native small-molecule networks of complete cells and potentially entire multi-cellular communities.
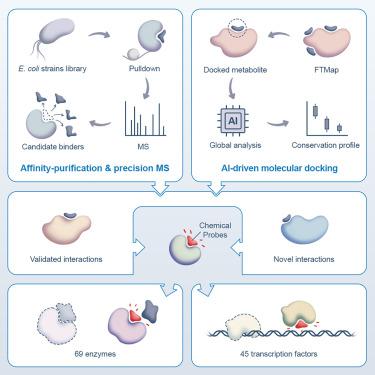
大肠杆菌中转录因子与必需酶的配体相互作用
蛋白质-代谢物相互作用的知识可以增强对生化过程的机制理解和化学探测,但内源性配体的发现仍然具有挑战性。在这里,我们结合了快速亲和纯化、精密质谱和高分辨率分子对接,精确绘制了296种化学上不同的小分子代谢物配体与69种不同的必需酶和45种转录因子的物理关联。然后,我们进行了系统的代谢途径整合、泛微生物进化预测和独立的深度生物物理表征实验,以确定配体界面的功能意义。这项工作揭示了在网络水平上控制功能串扰的原理,结合口袋守恒的不同模式,以及设计选择性化学探针的支架。这种结构上可分辨的配体相互作用体映射管道可以被缩放以照亮完整细胞和潜在的整个多细胞群落的天然小分子网络。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell
生物-生化与分子生物学
CiteScore
110.00
自引率
0.80%
发文量
396
审稿时长
2 months
期刊介绍:
Cells is an international, peer-reviewed, open access journal that focuses on cell biology, molecular biology, and biophysics. It is affiliated with several societies, including the Spanish Society for Biochemistry and Molecular Biology (SEBBM), Nordic Autophagy Society (NAS), Spanish Society of Hematology and Hemotherapy (SEHH), and Society for Regenerative Medicine (Russian Federation) (RPO).
The journal publishes research findings of significant importance in various areas of experimental biology, such as cell biology, molecular biology, neuroscience, immunology, virology, microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. The primary criterion for considering papers is whether the results contribute to significant conceptual advances or raise thought-provoking questions and hypotheses related to interesting and important biological inquiries.
In addition to primary research articles presented in four formats, Cells also features review and opinion articles in its "leading edge" section, discussing recent research advancements and topics of interest to its wide readership.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


