Leveraging a phased pangenome for haplotype design of hybrid potato
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
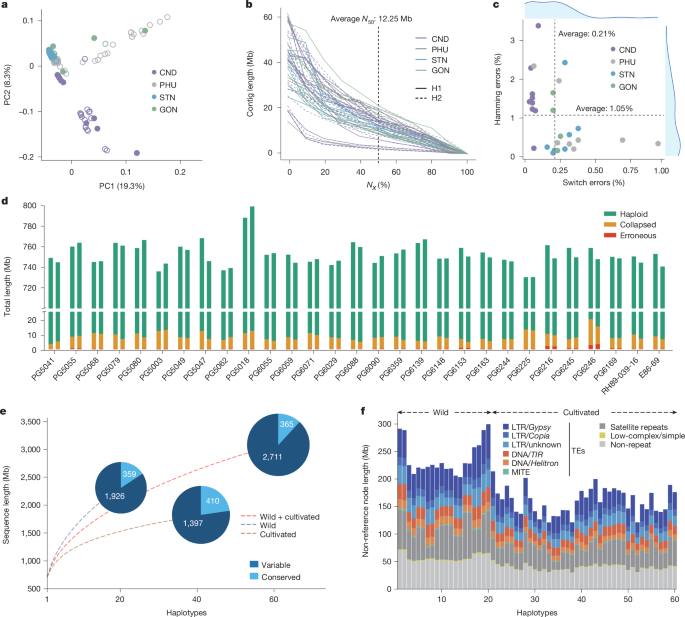
The tetraploid genome and clonal propagation of the cultivated potato (Solanum tuberosum L.)1,2 dictate a slow, non-accumulative breeding mode of the most important tuber crop. Transitioning potato breeding to a seed-propagated hybrid system based on diploid inbred lines has the potential to greatly accelerate its improvement3. Crucially, the development of inbred lines is impeded by manifold deleterious variants; explaining their nature and finding ways to eliminate them is the current focus of hybrid potato research4–10. However, most published diploid potato genomes are unphased, concealing crucial information on haplotype diversity and heterozygosity11–13. Here we develop a phased potato pangenome graph of 60 haplotypes from cultivated diploids and the ancestral wild species, and find evidence for the prevalence of transposable elements in generating structural variants. Compared with the linear reference, the graph pangenome represents a broader diversity (3,076 Mb versus 742 Mb). Notably, we observe enhanced heterozygosity in cultivated diploids compared with wild ones (14.0% versus 9.5%), indicating extensive hybridization during potato domestication. Using conservative criteria, we identify 19,625 putatively deleterious structural variants (dSVs) and reveal a biased accumulation of deleterious single nucleotide polymorphisms (dSNPs) around dSVs in coupling phase. Based on the graph pangenome, we computationally design ideal potato haplotypes with minimal dSNPs and dSVs. These advances provide critical insights into the genomic basis of clonal propagation and will guide breeders to develop a suite of promising inbred lines. A phased pangenome of potato constructed from 60 wild and cultivated haplotypes shows that substantial hybridization occurred during domestication and enables identification of many putative deleterious variants, providing a basis for the design of improved inbred lines.

利用分阶段泛基因组进行杂交马铃薯单倍型设计
栽培马铃薯(Solanum tuberosum L.)1,2的四倍体基因组和无性系繁殖决定了这种最重要的块茎作物的缓慢、非累积育种模式。将马铃薯育种转变为以二倍体自交系为基础的种子繁殖杂交系统,有可能大大加快马铃薯的改良。至关重要的是,自交系的发育受到多种有害变异的阻碍;解释它们的性质并找到消除它们的方法是杂交马铃薯目前研究的重点4,5,6,7,8,9,10。然而,大多数已发表的二倍体马铃薯基因组是未分期的,隐藏了单倍型多样性和杂合性的关键信息11,12,13。在这里,我们从栽培二倍体和祖先野生物种中建立了60个单倍型的阶段性马铃薯全基因组图,并找到了转座因子在产生结构变异中的普遍存在的证据。与线性参考相比,图形泛基因组表现出更广泛的多样性(3076 Mb比742 Mb)。值得注意的是,我们观察到栽培二倍体的杂合度比野生二倍体高(14.0%比9.5%),表明马铃薯驯化过程中存在广泛的杂交。使用保守标准,我们确定了19625个推定有害结构变异(dsv),并揭示了耦合期dsv周围有害单核苷酸多态性(dsnp)的偏倚积累。在泛基因组图谱的基础上,通过计算设计了具有最小dSNPs和最小dsv的马铃薯理想单倍型。这些进展为克隆繁殖的基因组基础提供了重要的见解,并将指导育种者开发一套有前途的自交系。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


