Correction to “Development of BromoTag: A “Bump-and-Hole”–PROTAC System To Induce Potent, Rapid, and Selective Degradation of Tagged Target Proteins”
IF 6.8
1区 医学
Q1 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
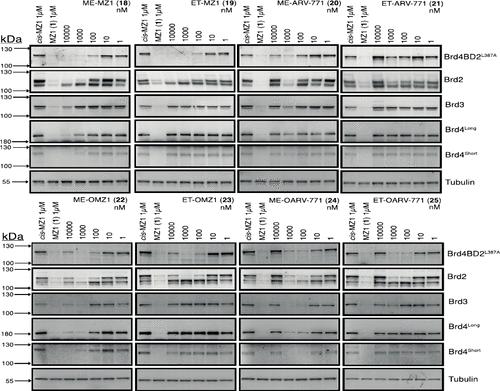
It has come to our attention that two of the Western blot panels in Figure 5, namely, the “Brd4 short” lanes for ET-OMZ1 (23) and ET-OARV-771 (25) were the same. This arose as a result of an honest mistake when choosing panels used from the corresponding uncropped blots. This is clear from the relevant Supplemental Figure (Figure S7, page 16 in the Supporting Information), which shows all the full-length uncropped blots that were used to prepare the final Figure 5. The figure has been corrected and shows the correct panel corresponding to “Brd4 short” for ET-OMZ1 (23). The panel corresponding to “Brd4 short” for compound ET-OARV-771 (25) was correct and thus did not require changes. Having recognized this issue, we decided to go over ALL the material again and found a few other minor issues. We discovered that the “Brd4 short” panel for ME-OARV-771 (24) was also incorrectly chosen from the corresponding uncropped blot (see the same relevant Supplemental Figure S7, on page 16 in the Supporting Information). This has now been corrected. We have also found that the “tubulin” panels for ET-MZ1 (19), ME-ARV-771 (20) and ET-ARV-771 (21) were incorrectly cropped to show 8 lanes instead of the 7 in question, keeping an additional irrelevant lane on the far right of the panels. We have now recropped these panels to remove this lane. Figure 5 with the corrected panels is shown above. The data analysis and conclusions are not affected by these mistakes. Page 15496 (middle of right column). The identity of the template backbone of the BromoTag vector donor was incorrectly quoted as “pcDNA5”. The correct vector is “pMK-RQ” as follows. HEK293 cells were transfected using a Fugene HD transfection reagent (Madison, Wisconsin, United States) or lipofectamine 2000 (Madison, Wisconsin, United States) simultaneously with three custom vectors including a px335 vector (Addgene) containing a U6-snRNA and Cas9D10A expression cassette, a pBABED vector (MRC PPU, Dundee University) harboring another U6-sgRNA and puromycin expression cassette, and finally a pMK-RQ vector containing an eGFP-P2A-BromoTag-Brd2 donor knock-in sequence. Page 15498 (bottom of left column). The size of the tissue culture plate used was incorrectly quoted as “100 cm”. The correct size is “10 cm” as follows. CRISPR-modified BromoTag-Brd2 HEK293 cells (5 × 106) were seeded on a 10 cm plate 24 h before treatment. We have revised the Supporting Information to add text to clarify the number of biological replicates (n) that were performed for each experiment. We have now also realized that the “Brd4 antibody” panel for PROTAC AGB1 (46) at the top right of Supplementary Figure 9 on page 24 in the Supporting Information (and the corresponding tubulin loading control) was from a different biological replicate of the same experiment than the one chosen in the corresponding main text figure (Figure 6, on page 15489). This has now been corrected in the Supporting Information. We also detected that the labels “cis-MZ1” and “MZ1 (1)” in Supplementary Figure 15 on page 27 in the Supporting Information were swapped accidentally. This has now been corrected in the Supporting Information. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.5c00108. DNA sequences of BromoTag degron components; junction PCR; genotyping of the span of eGFP-P2A-BromoTag in the heterozygous BromoTag-Brd2 Hek293 cell line; protein degradation profiles of PROTACs 18–25 for Brd4 long, Brd4 short, Brd3, Brd2, and Brd4BD2 L387A; protein degradation profiles of AGB1 (46), AGB2 (47), and AGB3 (48) for Brd4BD2 L387A and Brd2; original uncropped Western blot images; HPLC-HRMS traces for compounds 46–48 and 52; NMR spectra for compounds 46–48 and 52 (PDF) Correction to “Development of BromoTag: A “Bump-and-Hole”–PROTAC System To Induce Potent, Rapid, and Selective Degradation of Tagged Target Proteins” 2 views 0 shares 0 downloads Most electronic Supporting Information files are available without a subscription to ACS Web Editions. Such files may be downloaded by article for research use (if there is a public use license linked to the relevant article, that license may permit other uses). Permission may be obtained from ACS for other uses through requests via the RightsLink permission system: http://pubs.acs.org/page/copyright/permissions.html. This article has not yet been cited by other publications.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Medicinal Chemistry
医学-医药化学
CiteScore
4.00
自引率
11.00%
发文量
804
审稿时长
1.9 months
期刊介绍:
The Journal of Medicinal Chemistry is a prestigious biweekly peer-reviewed publication that focuses on the multifaceted field of medicinal chemistry. Since its inception in 1959 as the Journal of Medicinal and Pharmaceutical Chemistry, it has evolved to become a cornerstone in the dissemination of research findings related to the design, synthesis, and development of therapeutic agents.
The Journal of Medicinal Chemistry is recognized for its significant impact in the scientific community, as evidenced by its 2022 impact factor of 7.3. This metric reflects the journal's influence and the importance of its content in shaping the future of drug discovery and development. The journal serves as a vital resource for chemists, pharmacologists, and other researchers interested in the molecular mechanisms of drug action and the optimization of therapeutic compounds.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


