Structural variants of AcrIIC5 inhibit Cas9 via divergent binding interfaces
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
CRISPR-Cas is a bacterial defense system that employs RNA-guided endonucleases to destroy invading foreign nucleic acids. Bacteriophages produce anti-CRISPR (Acr) proteins to evade CRISPR-Cas defense during the infection. AcrIIC5, a type II-C Cas9 inhibitor, exhibits unusual variations in the local backbone fold between its orthologs. Here we investigated how the folding variations affect the inhibition of target Cas9 using AcrIIC5 orthologs. Structural comparison of free AcrIIC5Smu and AcrIIC5Nch confirmed that the folding variation correlated with characteristic indels in the helical region. Mutagenesis and biochemical assays combined with AlphaFold2 predictions identified key residues of AcrIIC5 orthologs important for Cas9 inhibition. Remarkably, AcrIIC5 orthologs employed divergent binding interfaces via folding variations to inhibit the Cas9 targets. Our study suggests that Acr proteins have evolved structural variants to diversify key interfaces for target Cas9, which could be beneficial for the adaptation of phages to evasive mutations on the Cas9 surface.
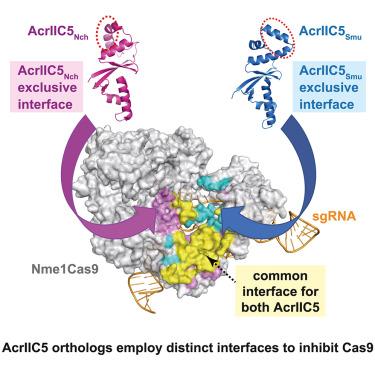
AcrIIC5的结构变体通过发散结合界面抑制Cas9
CRISPR-Cas是一种细菌防御系统,它利用rna引导的内切酶来破坏入侵的外来核酸。在感染过程中,噬菌体产生抗crispr (Acr)蛋白以逃避CRISPR-Cas的防御。II-C型Cas9抑制剂AcrIIC5在其同源物之间的局部骨干折叠中表现出不同寻常的变化。在这里,我们使用AcrIIC5同源物研究了折叠变化如何影响靶Cas9的抑制。自由的AcrIIC5Smu和AcrIIC5Nch的结构比较证实,折叠变化与螺旋区特征索引相关。诱变和生化分析结合AlphaFold2预测确定了对Cas9抑制重要的AcrIIC5同源物的关键残基。值得注意的是,AcrIIC5同源物通过折叠变化使用不同的结合界面来抑制Cas9靶标。我们的研究表明,Acr蛋白已经进化出结构变异,使靶Cas9的关键界面多样化,这可能有利于噬菌体适应Cas9表面的规避突变。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


