The regulatory landscape of 5′ UTRs in translational control during zebrafish embryogenesis
IF 10.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
The 5′ UTRs of mRNAs are critical for translation regulation during development, but their in vivo regulatory features are poorly characterized. Here, we report the regulatory landscape of 5′ UTRs during early zebrafish embryogenesis using a massively parallel reporter assay of 18,154 sequences coupled to polysome profiling. We found that the 5′ UTR suffices to confer temporal dynamics to translation initiation and identified 86 motifs enriched in 5′ UTRs with distinct ribosome recruitment capabilities. A quantitative deep learning model, Danio Optimus 5-Prime (DaniO5P), identified a combined role for 5′ UTR length, translation initiation site context, upstream AUGs, and sequence motifs on ribosome recruitment. DaniO5P predicts the activities of maternal and zygotic 5′ UTR isoforms and indicates that modulating 5′ UTR length and motif grammar contributes to translation initiation dynamics. This study provides a first quantitative model of 5′ UTR-based translation regulation in development and lays the foundation for identifying the underlying molecular effectors.
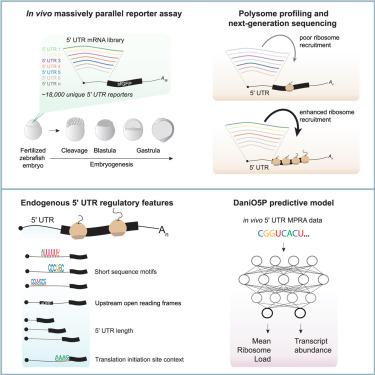
斑马鱼胚胎发生过程中5 ' utr在翻译控制中的调控前景
mrna的5 ' utr在发育过程中对翻译调控至关重要,但其体内调控特征尚不清楚。在这里,我们报告了5 ' utr在早期斑马鱼胚胎发生过程中的调控景观,使用了18154个序列与多体分析相结合的大规模平行报告分析。我们发现5 ' UTR足以赋予翻译起始时间动力学,并鉴定出86个富含5 ' UTR的基序,它们具有不同的核糖体招募能力。定量深度学习模型Danio Optimus 5- prime (DaniO5P)确定了5 ' UTR长度、翻译起始位点上下文、上游aug和序列基序在核糖体招募中的综合作用。DaniO5P可以预测母体和受精卵5 ' UTR同工型的活性,并表明调节5 ' UTR长度和基序语法有助于翻译起始动力学。本研究提供了第一个基于5 ' utr的翻译调控在发育中的定量模型,为确定潜在的分子效应奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


