Defining proteoform-specific interactions for drug targeting in a native cell signalling environment
IF 19.2
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
Understanding the dynamics of membrane protein–ligand interactions within a native lipid bilayer is a major goal for drug discovery. Typically, cell-based assays are used, however, they are often blind to the effects of protein modifications. In this study, using the archetypal G protein-coupled receptor rhodopsin, we found that the receptor and its effectors can be released directly from retina rod disc membranes using infrared irradiation in a mass spectrometer. Subsequent isolation and dissociation by infrared multiphoton dissociation enabled the sequencing of individual retina proteoforms. Specifically, we categorized distinct proteoforms of rhodopsin, localized labile palmitoylations, discovered a Gβγ proteoform that abolishes membrane association and defined lipid modifications on G proteins that influence their assembly. Given reports of undesirable side-effects involving vision, we characterized the off-target drug binding of two phosphodiesterase 5 inhibitors, vardenafil and sildenafil, to the retina rod phosphodiesterase 6 (PDE6). The results demonstrate differential off-target reactivity with PDE6 and an interaction preference for lipidated proteoforms of G proteins. In summary, this study highlights the opportunities for probing proteoform–ligand interactions within natural membrane environments. G protein-coupled receptors and their effectors can now be released directly from a lipid bilayer using infrared irradiation for proteoform-level characterization by native top-down mass spectrometry. This represents a critical development for drug discovery, as the direct role of post-translational modifications in protein–protein and protein–drug interactions can be characterized.

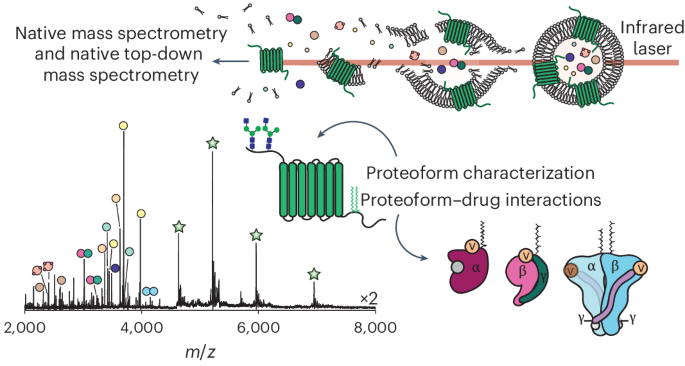
在天然细胞信号环境中定义药物靶向的蛋白形式特异性相互作用
了解天然脂质双分子层内膜蛋白-配体相互作用的动力学是药物发现的主要目标。通常使用的是基于细胞的测定,然而,它们往往对蛋白质修饰的影响视而不见。在本研究中,我们使用原型G蛋白偶联受体视紫红质,我们发现受体及其效应器可以直接从视网膜棒盘膜上释放。随后通过红外多光子解离进行分离和解离,可以对单个视网膜蛋白形态进行测序。具体来说,我们分类了不同的视紫红质蛋白形式,局部不稳定的棕榈酰化,发现了一种Gβγ蛋白形式,它可以消除膜结合,并定义了影响G蛋白组装的脂质修饰。鉴于涉及视力的不良副作用的报告,我们描述了两种磷酸二酯酶5抑制剂伐地那非和西地那非与视网膜棒磷酸二酯酶6 (PDE6)的脱靶药物结合。结果表明,PDE6具有不同的脱靶反应性,G蛋白的脂化蛋白形式具有相互作用偏好。总之,这项研究强调了在天然膜环境中探测蛋白质-配体相互作用的机会。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature chemistry
化学-化学综合
CiteScore
29.60
自引率
1.40%
发文量
226
审稿时长
1.7 months
期刊介绍:
Nature Chemistry is a monthly journal that publishes groundbreaking and significant research in all areas of chemistry. It covers traditional subjects such as analytical, inorganic, organic, and physical chemistry, as well as a wide range of other topics including catalysis, computational and theoretical chemistry, and environmental chemistry.
The journal also features interdisciplinary research at the interface of chemistry with biology, materials science, nanotechnology, and physics. Manuscripts detailing such multidisciplinary work are encouraged, as long as the central theme pertains to chemistry.
Aside from primary research, Nature Chemistry publishes review articles, news and views, research highlights from other journals, commentaries, book reviews, correspondence, and analysis of the broader chemical landscape. It also addresses crucial issues related to education, funding, policy, intellectual property, and the societal impact of chemistry.
Nature Chemistry is dedicated to ensuring the highest standards of original research through a fair and rigorous review process. It offers authors maximum visibility for their papers, access to a broad readership, exceptional copy editing and production standards, rapid publication, and independence from academic societies and other vested interests.
Overall, Nature Chemistry aims to be the authoritative voice of the global chemical community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


