Cell state-dependent allelic effects and contextual Mendelian randomization analysis for human brain phenotypes
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
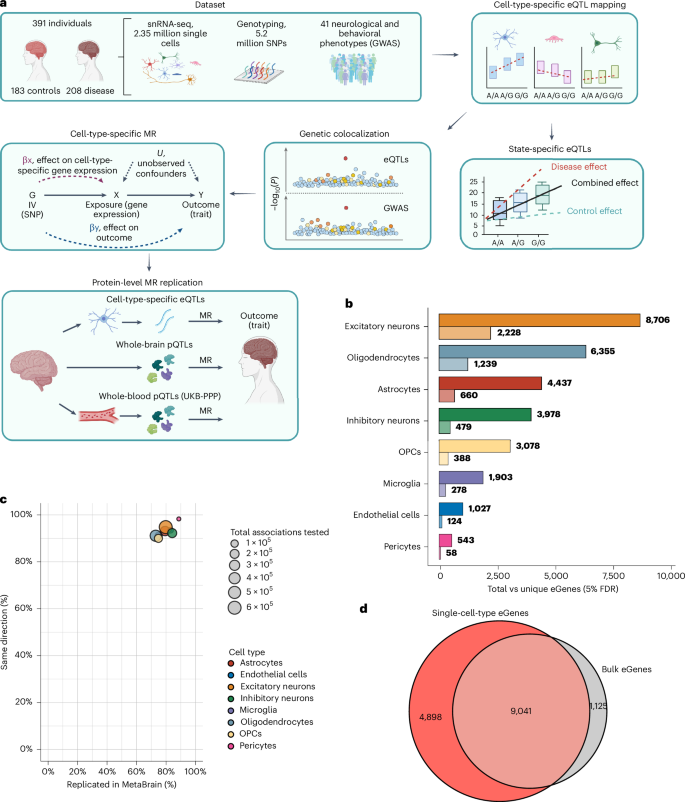
Gene expression quantitative trait loci are widely used to infer relationships between genes and central nervous system (CNS) phenotypes; however, the effect of brain disease on these inferences is unclear. Using 2,348,438 single-nuclei profiles from 391 disease-case and control brains, we report 13,939 genes whose expression correlated with genetic variation, of which 16.7–40.8% (depending on cell type) showed disease-dependent allelic effects. Across 501 colocalizations for 30 CNS traits, 23.6% had a disease dependency, even after adjusting for disease status. To estimate the unconfounded effect of genes on outcomes, we repeated the analysis using nondiseased brains (n = 183) and reported an additional 91 colocalizations not present in the larger mixed disease and control dataset, demonstrating enhanced interpretation of disease-associated variants. Principled implementation of single-cell Mendelian randomization in control-only brains identified 140 putatively causal gene–trait associations, of which 11 were replicated in the UK Biobank, prioritizing candidate peripheral biomarkers predictive of CNS outcomes. Analysis of single nuclei RNA-sequencing data across eight major brain cell types identifies putative causal associations between cell-type-specific expression and brain phenotypes.

人类大脑表型的细胞状态依赖性等位基因效应和上下文孟德尔随机化分析
基因表达数量性状位点被广泛用于推断基因与中枢神经系统表型之间的关系;然而,脑部疾病对这些推论的影响尚不清楚。利用来自391个疾病病例和对照大脑的2,348,438个单核谱,我们报告了13,939个基因的表达与遗传变异相关,其中16.7-40.8%(取决于细胞类型)表现出疾病依赖性等位基因效应。在30个中枢神经系统特征的501个共定位中,即使在调整疾病状态后,23.6%的人仍存在疾病依赖性。为了估计基因对结果的明确影响,我们使用未患病的大脑(n = 183)重复了分析,并报告了在更大的混合疾病和对照数据集中不存在的另外91个共定位,证明了对疾病相关变异的增强解释。在对照组中,单细胞孟德尔随机化的原则实施确定了140个推定的因果基因性状关联,其中11个在英国生物银行得到了重复,优先考虑了预测中枢神经系统预后的候选外周生物标志物。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


