DYRK1A-TGF-β signaling axis determines sensitivity to OXPHOS inhibition in hepatocellular carcinoma
IF 10.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Intervening in mitochondrial oxidative phosphorylation (OXPHOS) has emerged as a potential therapeutic strategy for certain types of cancers. Employing kinome-based CRISPR screen, we find that knockout of dual-specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) synergizes with OXPHOS inhibitor IACS-010759 in liver cancer cells. Targeting DYRK1A combined with OXPHOS inhibitors activates TGF-β signaling, which is crucial for OXPHOS-inhibition-triggered cell death. Mechanistically, upregulation of glutamine transporter solute carrier family 1 member 5 (SLC1A5) transcription compensates for the increased glutamine requirement upon OXPHOS inhibition. DYRK1A directly phosphorylates SMAD3 Thr132, thereby suppressing the negative impact of TGF-β signaling on transcription of SLC1A5, leading to intrinsic resistance of liver cancer cells to OXPHOS inhibition. Moreover, we demonstrate the therapeutic efficacy of IACS-010759 in combination with DYRK1A inhibition in multiple liver cancer models, including xenografts, patient-derived xenografts, and spontaneous tumor model. Our study elucidates how the DYRK1A-TGF-β signaling axis controls the response of tumor cells to OXPHOS inhibition and provides valuable insights into targeting OXPHOS for liver cancer therapy.
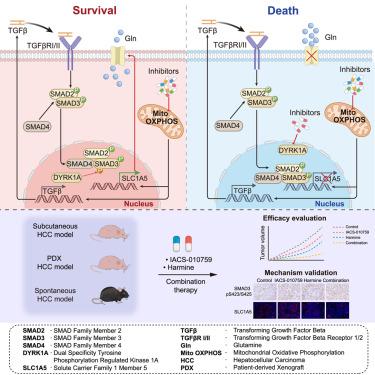
DYRK1A-TGF-β信号轴决定肝癌对OXPHOS抑制的敏感性
干预线粒体氧化磷酸化(OXPHOS)已成为某些类型癌症的潜在治疗策略。通过基于kinomin的CRISPR筛选,我们发现在肝癌细胞中敲除双特异性酪氨酸磷酸化调节激酶1A (DYRK1A)与OXPHOS抑制剂IACS-010759协同作用。靶向DYRK1A与OXPHOS抑制剂联合激活TGF-β信号,这对于OXPHOS抑制剂引发的细胞死亡至关重要。机制上,谷氨酰胺转运体溶质载体家族1成员5 (SLC1A5)转录的上调补偿了OXPHOS抑制时谷氨酰胺需求的增加。DYRK1A直接磷酸化SMAD3 Thr132,从而抑制TGF-β信号传导对SLC1A5转录的负面影响,导致肝癌细胞对OXPHOS抑制的内在抗性。此外,我们证明了IACS-010759联合DYRK1A抑制在多种肝癌模型中的治疗效果,包括异种移植、患者来源的异种移植和自发肿瘤模型。我们的研究阐明了DYRK1A-TGF-β信号轴如何控制肿瘤细胞对OXPHOS抑制的反应,并为靶向OXPHOS治疗肝癌提供了有价值的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


