Interpretable identification of cancer genes across biological networks via transformer-powered graph representation learning
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
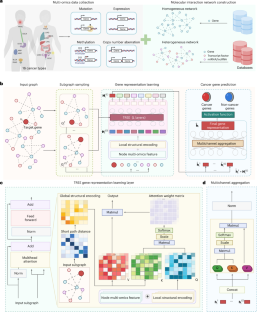
Graph representation learning has been leveraged to identify cancer genes from biological networks. However, its applicability is limited by insufficient interpretability and generalizability under integrative network analysis. Here we report the development of an interpretable and generalizable transformer-based model that accurately predicts cancer genes by leveraging graph representation learning and the integration of multi-omics data with the topologies of homogeneous and heterogeneous networks of biological interactions. The model allows for the interpretation of the respective importance of multi-omic and higher-order structural features, achieved state-of-the-art performance in the prediction of cancer genes across biological networks (including networks of interactions between miRNA and proteins, transcription factors and proteins, and transcription factors and miRNA) in pan-cancer and cancer-specific scenarios, and predicted 57 cancer-gene candidates (including three genes that had not been identified by other models) among 4,729 unlabelled genes across 8 pan-cancer datasets. The model’s interpretability and generalization may facilitate the understanding of gene-related regulatory mechanisms and the discovery of new cancer genes. An interpretable transformer-based model leveraging graph representation learning accurately predicts cancer genes across homogeneous and heterogeneous pan-cancer networks of biological interactions.

通过变压器驱动的图表示学习跨生物网络的可解释的癌症基因鉴定
图表示学习已被用于从生物网络中识别癌症基因。但在综合网络分析下,其可解释性和概括性不足,限制了其适用性。在这里,我们报告了一种可解释和可推广的基于转换器的模型的发展,该模型通过利用图表示学习和多组学数据与同质和异质生物相互作用网络拓扑结构的集成来准确预测癌症基因。该模型允许解释多组学和高阶结构特征各自的重要性,在泛癌症和癌症特定情况下,在跨生物网络(包括miRNA与蛋白质、转录因子与蛋白质、转录因子与miRNA之间的相互作用网络)预测癌症基因方面取得了最先进的性能。并在8个泛癌症数据集中的4729个未标记基因中预测了57个候选癌症基因(包括3个未被其他模型识别的基因)。该模型的可解释性和泛化性可能有助于理解基因相关调控机制和发现新的癌症基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


