Structural polymorphism and diversity of human segmental duplications
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
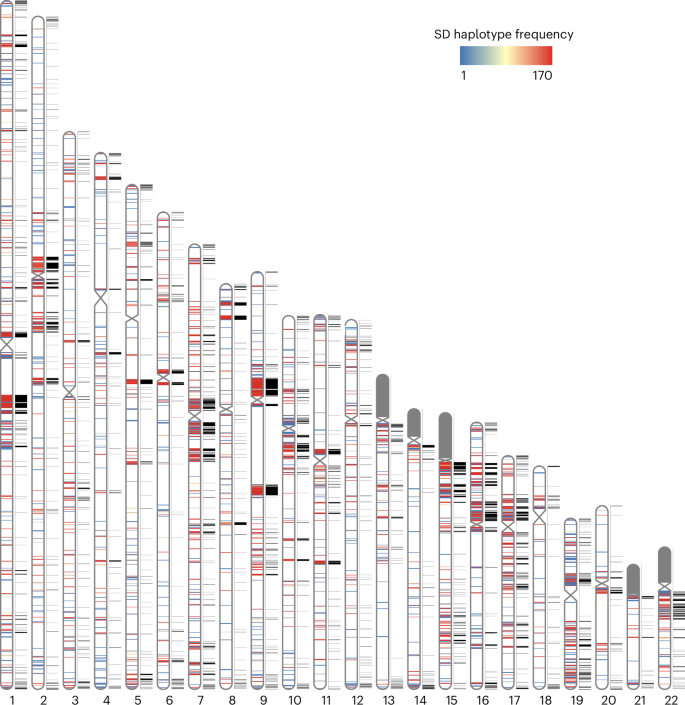
Segmental duplications (SDs) contribute significantly to human disease, evolution and diversity but have been difficult to resolve at the sequence level. We present a population genetics survey of SDs by analyzing 170 human genome assemblies (from 85 samples representing 38 Africans and 47 non-Africans) in which the majority of autosomal SDs are fully resolved using long-read sequence assembly. Excluding the acrocentric short arms and sex chromosomes, we identify 173.2 Mb of duplicated sequence (47.4 Mb not present in the telomere-to-telomere reference) distinguishing fixed from structurally polymorphic events. We find that intrachromosomal SDs are among the most variable, with rare events mapping near their progenitor sequences. African genomes harbor significantly more intrachromosomal SDs and are more likely to have recently duplicated gene families with higher copy numbers than non-African samples. Comparison to a resource of 563 million full-length isoform sequencing reads identifies 201 novel, potentially protein-coding genes corresponding to these copy number polymorphic SDs. Analysis of 170 human genomes assembled using long-read sequencing provides a map of structural variation within regions of segmental duplication and identifies novel candidate protein-coding genes supported by full-length Iso-Seq reads.

人类片段复制的结构多态性和多样性
片段重复(SDs)对人类疾病、进化和多样性有重要贡献,但难以在序列水平上解决。我们通过分析170个人类基因组组合(来自85个样本,代表38个非洲人和47个非非洲人),对SDs进行了群体遗传学调查,其中大多数常染色体SDs使用长读序列组装完全解决。排除顶心短臂和性染色体,我们鉴定了173.2 Mb的重复序列(47.4 Mb不存在于端粒到端粒的参考中),以区分固定和结构多态性事件。我们发现染色体内的SDs是最可变的,罕见的事件映射在它们的祖序列附近。与非非洲样本相比,非洲基因组含有更多的染色体内SDs,并且更有可能具有较高拷贝数的近期重复基因家族。与5.63亿个全长同种异构体测序的资源相比,鉴定出201个新的、潜在的蛋白质编码基因,与这些拷贝数多态性SDs相对应。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


