Refining breast cancer genetic risk and biology through multi-ancestry fine-mapping analyses of 192 risk regions
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Genome-wide association studies have identified approximately 200 genetic risk loci for breast cancer, but the causal variants and target genes are mostly unknown. We sought to fine-map all known breast cancer risk loci using genome-wide association study data from 172,737 female breast cancer cases and 242,009 controls of African, Asian and European ancestry. We identified 332 independent association signals for breast cancer risk, including 131 signals not reported previously, and for 50 of them, we narrowed the credible causal variants down to a single variant. Analyses integrating functional genomics data identified 195 putative susceptibility genes, enriched in PI3K/AKT, TNF/NF-κB, p53 and Wnt/β-catenin pathways. Single-cell RNA sequencing or in vitro experiment data provided additional functional evidence for 105 genes. Our study uncovered large numbers of association signals and candidate susceptibility genes for breast cancer, uncovered breast cancer genetics and biology, and supported the value of including multi-ancestry data in fine-mapping analyses. Multi-ancestry fine-mapping of breast cancer susceptibility regions identifies candidate causal variants and prioritizes likely effector genes supported by functional genomic evidence.
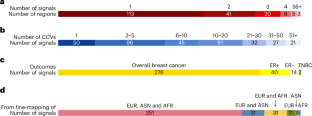

通过对192个风险区域的多祖先精细图谱分析,提炼乳腺癌遗传风险和生物学
全基因组关联研究已经确定了大约200个乳腺癌的遗传风险位点,但其因果变异和靶基因大多是未知的。我们利用来自172737例女性乳腺癌病例和242009例非洲、亚洲和欧洲血统对照的全基因组关联研究数据,试图精细绘制所有已知的乳腺癌风险位点。我们确定了332个与乳腺癌风险相关的独立信号,包括131个以前未报道的信号,其中50个信号,我们将可信的因果变异缩小到一个变异。整合功能基因组学数据的分析确定了195个推测的易感基因,这些基因在PI3K/AKT、TNF/NF-κB、p53和Wnt/β-catenin通路中富集。单细胞RNA测序或体外实验数据为105个基因提供了额外的功能证据。我们的研究揭示了乳腺癌的大量关联信号和候选易感基因,揭示了乳腺癌的遗传学和生物学,支持了多祖先数据在精细图谱分析中的价值。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


