The highly allo-autopolyploid modern sugarcane genome and very recent allopolyploidization in Saccharum
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
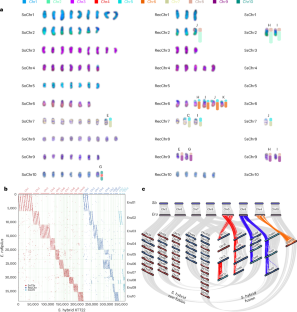
Modern sugarcane, a highly allo-autopolyploid organism, has a very complex genome. In the present study, the karyotype and genome architecture of modern sugarcane were investigated, resulting in a genome assembly of 97 chromosomes (8.84 Gb). The allopolyploid genome was divided into subgenomes from Saccharum officinarum (Soh) and S. spontaneum (Ssh), with Soh dominance in the Saccharum hybrid (S. hybrid). Genome shock affected transcriptome dynamics during allopolyploidization. Analysis of an inbreeding population with 192 individuals revealed the underlying genetic basis of transgressive segregation. Population genomics of 310 Saccharum accessions clarified the breeding history of modern sugarcane. Using the haplotype-resolved S. hybrid genome as a reference, genome-wide association studies identified a potential candidate gene for sugar content from S. spontaneum. These findings illuminate the complex genome evolution of allopolyploids, offering opportunities for genomic enhancements and innovative breeding strategies for sugarcane. A haplotype-resolved genome of hybrid sugarcane cultivar XTT22 and population analyses of Saccharum accessions highlight the genome evolution of allopolyploids and provide opportunities for sugarcane breeding.

现代甘蔗的高度异源自多倍体基因组和最近的甘蔗异源多倍体
现代甘蔗是一种高度同源多倍体生物,具有非常复杂的基因组。本研究对现代甘蔗的核型和基因组结构进行了研究,得到了97条染色体(8.84 Gb)的基因组组装。结果表明,Saccharum officinarum (Soh)和S. spontaneum (Ssh)的异源多倍体基因组分为两个亚基因组,其中Soh在Saccharum杂种(S. hybrid)中占优势。基因组冲击影响了异源多倍体化过程中的转录组动力学。对一个有192个个体的近交群体的分析揭示了越界分离的潜在遗传基础。310份Saccharum材料的群体基因组学分析阐明了现代甘蔗的育种历史。利用单倍型分离的葡萄杂交基因组作为参考,全基因组关联研究确定了一个潜在的候选基因。这些发现阐明了异源多倍体复杂的基因组进化,为甘蔗基因组增强和创新育种策略提供了机会。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


