Structural insights into the biochemical mechanism of the E2/E3 hybrid enzyme UBE2O
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The E2/E3 hybrid enzyme UBE2O plays important roles in key biological events, but its autoubiquitination mechanism remains largely unclear. In this study, we determined the crystal structures of full-length (FL) UBE2O from Trametes pubescens (tp) and its ubiquitin-conjugating (UBC) domain. The dimeric FL-tpUBE2O structure revealed interdomain interactions between the conserved regions (CR1-CR2) and UBC. The dimeric intermolecular and canonical ubiquitin/UBC interactions are mechanistically important for UBE2O functions in catalyzing the formation of free polyubiquitin chains and substrate ubiquitination. Beyond dimerization, autoubiquitination within the CR1-CR2 domain also regulates tpUBE2O activity. Additionally, we show that tpUBE2O catalyzes the formation of all seven types of polyubiquitin chains in vitro. The CR1-CR2/UBC and canonical ubiquitin/UBC interactions are important for the polyubiquitination of AMP-activated protein kinase α2 (AMPKα2) by human UBE2O (hUBE2O), which leads to tumorigenesis. These structural insights lay the groundwork for understanding UBE2O’s mechanisms and developing structure-based therapeutics targeting UBE2O.
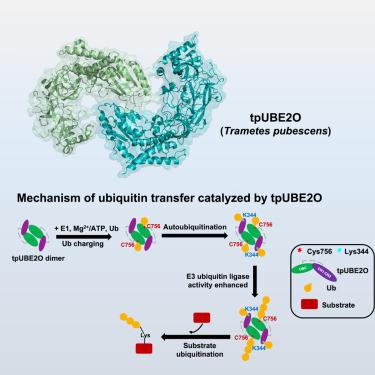
从结构上揭示 E2/E3 混合酶 UBE2O 的生化机制
E2/E3杂交酶UBE2O在关键生物学事件中发挥重要作用,但其自泛素化机制尚不清楚。在这项研究中,我们确定了从Trametes pubescens (tp)中提取的全长(FL) ube20及其泛素偶联(UBC)结构域的晶体结构。二聚体FL-tpUBE2O结构揭示了保守区域(CR1-CR2)和UBC之间的域间相互作用。二聚体分子间和典型泛素/UBC相互作用对ube20催化游离多泛素链的形成和底物泛素化的功能具有重要的机制意义。除了二聚化外,CR1-CR2结构域内的自泛素化也调节tpUBE2O活性。此外,我们发现tpube20在体外催化所有七种多泛素链的形成。CR1-CR2/UBC和典型泛素/UBC相互作用对于人UBE2O (hUBE2O)对amp活化的蛋白激酶α2 (AMPKα2)的多泛素化至关重要,从而导致肿瘤的发生。这些结构见解为理解UBE2O的机制和开发针对UBE2O的基于结构的治疗方法奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


