The structures of the peptide transporters SLC15A3 and SLC15A4 reveal the recognition mechanisms for substrate and TASL
IF 4.4
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
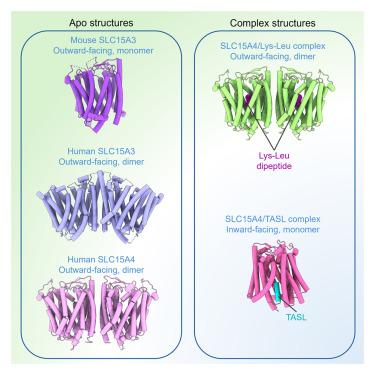
The solute carrier family 15 members 3 and 4 (SLC15A3 and SLC15A4) are closely related endolysosomal peptide transporters that transport free histidine and certain dipeptides from the lumen to cytosol. Besides, SLC15A4 also functions as a scaffold protein for the recruitment of the adapter TASL for interferon regulatory factor 5 (IRF5) activation downstream of innate immune TLR7-9 signaling. However, the molecular basis for the substrate recognition and TASL recruitment by these membrane proteins is not well understood. Here, we report the cryoelectron microscopy (cryo-EM) structure of apo SLC15A3 and structures of SLC15A4 in the absence or presence of the substrate, revealing the specific dipeptide recognition mechanism. Each SLC15A3 and SLC15A4 protomer adopts an outward-facing conformation. Furthermore, we also present the cryo-EM structure of a SLC15A4-TASL complex. The N terminal region of TASL forms a helical structure that inserts deeply into the inward-facing cavity of SLC15A4.
肽转运体SLC15A3和SLC15A4的结构揭示了对底物和TASL的识别机制
溶质载体家族15成员3和4 (SLC15A3和SLC15A4)是密切相关的内溶酶体肽转运体,将游离组氨酸和某些二肽从管腔运输到细胞质。此外,SLC15A4还作为支架蛋白,在先天免疫TLR7-9信号下游募集适配体TASL,激活干扰素调节因子5 (IRF5)。然而,这些膜蛋白识别底物和募集TASL的分子基础尚不清楚。在此,我们报道了载脂蛋白SLC15A3和SLC15A4在无底物或存在底物情况下的低温电镜结构,揭示了特定的二肽识别机制。每个SLC15A3和SLC15A4原聚体采用外向构象。此外,我们还展示了SLC15A4-TASL配合物的低温电镜结构。TASL的N端区域形成螺旋状结构,深入SLC15A4的内腔。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Structure
生物-生化与分子生物学
CiteScore
8.90
自引率
1.80%
发文量
155
审稿时长
3-8 weeks
期刊介绍:
Structure aims to publish papers of exceptional interest in the field of structural biology. The journal strives to be essential reading for structural biologists, as well as biologists and biochemists that are interested in macromolecular structure and function. Structure strongly encourages the submission of manuscripts that present structural and molecular insights into biological function and mechanism. Other reports that address fundamental questions in structural biology, such as structure-based examinations of protein evolution, folding, and/or design, will also be considered. We will consider the application of any method, experimental or computational, at high or low resolution, to conduct structural investigations, as long as the method is appropriate for the biological, functional, and mechanistic question(s) being addressed. Likewise, reports describing single-molecule analysis of biological mechanisms are welcome.
In general, the editors encourage submission of experimental structural studies that are enriched by an analysis of structure-activity relationships and will not consider studies that solely report structural information unless the structure or analysis is of exceptional and broad interest. Studies reporting only homology models, de novo models, or molecular dynamics simulations are also discouraged unless the models are informed by or validated by novel experimental data; rationalization of a large body of existing experimental evidence and making testable predictions based on a model or simulation is often not considered sufficient.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


