An amino acid-resolution interactome for motile cilia identifies the structure and function of ciliopathy protein complexes
IF 8.7
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Motile cilia are ancient, evolutionarily conserved organelles whose dysfunction underlies motile ciliopathies, a broad class of human diseases. Motile cilia contain a myriad of different proteins that assemble into an array of distinct machines, and understanding the interactions and functional hierarchies among them presents an important challenge. Here, we defined the protein interactome of motile axonemes using cross-linking mass spectrometry in Tetrahymena thermophila. From over 19,000 cross-links, we identified over 4,700 unique amino acid interactions among over 1,100 distinct proteins, providing both macromolecular and atomic-scale insights into diverse ciliary machines, including the intraflagellar transport system, axonemal dynein arms, radial spokes, the 96-nm ruler, and microtubule inner proteins. Guided by this dataset, we used vertebrate multiciliated cells to reveal functional interactions among several poorly defined human ciliopathy proteins. This dataset provides a resource for studying the biology of an ancient organelle and the molecular etiology of human genetic disease.
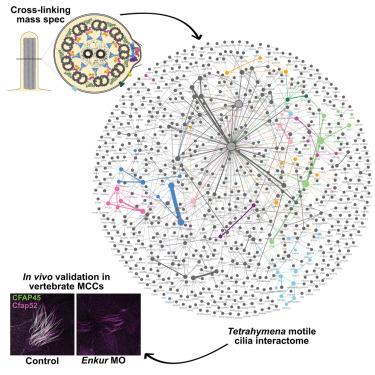
运动纤毛的氨基酸分辨率相互作用组识别纤毛病蛋白复合物的结构和功能
运动性纤毛是一种古老的、进化上保守的细胞器,其功能障碍是运动性纤毛病(一类广泛的人类疾病)的基础。运动纤毛包含无数不同的蛋白质,这些蛋白质组装成一系列不同的机器,了解它们之间的相互作用和功能层次是一个重要的挑战。在这里,我们用交联质谱法定义了嗜热四膜虫运动轴突的蛋白质相互作用组。从19,000多个交联中,我们确定了超过1,100种不同蛋白质之间的4,700多种独特氨基酸相互作用,为各种纤毛机器提供了大分子和原子尺度的见解,包括鞭毛内运输系统,轴突动力蛋白臂,径向辐条,96纳米尺子和微管内部蛋白质。在此数据集的指导下,我们使用脊椎动物多纤毛细胞来揭示几种定义不清的人类纤毛病蛋白之间的功能相互作用。该数据集为研究古代细胞器的生物学和人类遗传疾病的分子病因学提供了资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Developmental cell
生物-发育生物学
CiteScore
18.90
自引率
1.70%
发文量
203
审稿时长
3-6 weeks
期刊介绍:
Developmental Cell, established in 2001, is a comprehensive journal that explores a wide range of topics in cell and developmental biology. Our publication encompasses work across various disciplines within biology, with a particular emphasis on investigating the intersections between cell biology, developmental biology, and other related fields. Our primary objective is to present research conducted through a cell biological perspective, addressing the essential mechanisms governing cell function, cellular interactions, and responses to the environment. Moreover, we focus on understanding the collective behavior of cells, culminating in the formation of tissues, organs, and whole organisms, while also investigating the consequences of any malfunctions in these intricate processes.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


