Ignoring population structure in hominin evolutionary models can lead to the inference of spurious admixture events
IF 13.9
1区 生物学
Q1 ECOLOGY
引用次数: 0
Abstract
Genomic and ancient DNA data have revolutionized palaeoanthropology and our vision of human evolution, with indisputable landmarks like the sequencing of Neanderthal and Denisovan genomes. Yet, using genetic data to identify, date and quantify evolutionary events—such as ancient bottlenecks or admixture—is not straightforward, as inferences may depend on model assumptions. In the last two decades, the idea that Neanderthals and members of the Homo sapiens lineage interbred has gained momentum. From the status of unlikely theory, it has reached consensus among human evolutionary biologists. This theory is mainly supported by statistical approaches that depend on demographic models minimizing or ignoring population structure, despite its widespread occurrence and the fact that, when ignored, population structure can lead to the inference of spurious demographic events. We simulated genomic data under a structured and admixture-free model of human evolution, and found that all the tested admixture approaches identified long Neanderthal fragments in our simulated genomes and an admixture event that never took place. We also observed that several published admixture models failed to predict important empirical diversity or admixture statistics, and that we could identify several scenarios from our structured model that better predicted these statistics jointly. Using a simulated time series of ancient DNA, the structured scenarios could also predict the trajectory of the empirical D statistics. Our results suggest that models accounting for population structure are fundamental to improve our understanding of human evolution, and that admixture between Neanderthals and H. sapiens needs to be re-evaluated in the light of structured models. Beyond the Neanderthal case, we argue that ancient hybridization events, which are increasingly documented in many species, including with other hominins, may also benefit from such re-evaluation. Simulating a metapopulation of human evolution without Neanderthal introgression into Homo sapiens still identifies Neanderthal fragments in simulated genomes, and an admixture event that never took place. This indicates that population structure must be accounted for in human evolutionary genomics and that putative ancient hybridization events should be reinterpreted in this light.

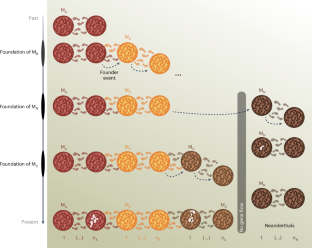
在类人进化模型中忽略种群结构会导致推断出虚假的混杂事件
基因组和古 DNA 数据彻底改变了古人类学和我们对人类进化的认识,尼安德特人和丹尼索瓦人基因组测序等里程碑事件无可争议。然而,利用基因数据来识别、确定进化事件(如远古瓶颈或混杂)的时间和数量并不简单,因为推论可能取决于模型假设。在过去的二十年里,尼安德特人与智人血统成员杂交的观点得到了越来越多的支持。它从一种不太可能的理论,发展成为人类进化生物学家的共识。支持这一理论的主要是统计方法,这些方法依赖于最小化或忽略种群结构的人口统计模型,尽管种群结构广泛存在,而且忽略种群结构会导致推断出虚假的人口统计事件。我们在一个结构化和无混杂的人类进化模型下模拟了基因组数据,发现所有测试过的混杂方法都在我们模拟的基因组中发现了长尼安德特人片段,以及从未发生过的混杂事件。我们还观察到,一些已发表的混杂模型未能预测重要的经验多样性或混杂统计数据,而我们可以从我们的结构化模型中找出几种情况,更好地共同预测这些统计数据。利用古 DNA 的模拟时间序列,结构化方案还能预测经验 D 统计量的轨迹。我们的研究结果表明,考虑种群结构的模型对于提高我们对人类进化的理解至关重要,尼安德特人与智人之间的混杂需要根据结构化模型重新评估。除了尼安德特人之外,我们还认为,在许多物种(包括与其他类人猿)中被越来越多地记录下来的古代杂交事件也可能从这种重新评估中受益。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature ecology & evolution
Agricultural and Biological Sciences-Ecology, Evolution, Behavior and Systematics
CiteScore
22.20
自引率
2.40%
发文量
282
期刊介绍:
Nature Ecology & Evolution is interested in the full spectrum of ecological and evolutionary biology, encompassing approaches at the molecular, organismal, population, community and ecosystem levels, as well as relevant parts of the social sciences. Nature Ecology & Evolution provides a place where all researchers and policymakers interested in all aspects of life's diversity can come together to learn about the most accomplished and significant advances in the field and to discuss topical issues. An online-only monthly journal, our broad scope ensures that the research published reaches the widest possible audience of scientists.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


