Backbone resonance assignments of the C-terminal thioesterase domain of tyrocidine synthetase C
Abstract
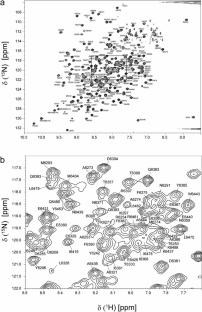
Natural macrocyclic peptides produced by microorganisms serve as valuable resources for therapeutic compounds, including antibiotics, anticancer agents, and immune suppressive agents. Nonribosomal peptide synthetases (NRPSs) are responsible for the biosynthesis of macrocyclic peptides. NRPSs are large multimodular enzymes, and each module recognizes and incorporates one specific amino acid into the polypeptide product. In the final biosynthetic step, the mature linear peptide precursor is subject to head-to-tail cyclization by the thioesterase (TE) domain in the C-terminal module. Since the TE domains can autonomously catalyze the cyclization of diverse linear peptide substrates, isolated TE domains can be used to produce natural product derivatives. To understand the mechanism of TE domains in NRPSs as a base for therapeutic applications, we investigated the TE domain (residues 6236–6486) of tyrocidine synthetase TycC by NMR. Tyrocidine is a cyclic decapeptide with antibiotic activity, and TycC-TE catalyzes the cyclization of the linear decapeptide precursor. Here, we report the backbone resonance assignments of TycC-TE. The assignments of TycC-TE provide the basis for NMR investigations of the structure and substrate-recognition mode of the TE domain in NRPS.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


