Bifunctional Probe Propelling Multipath Strand Displacement Amplification Tandem CRISPR/Cas12a for Ultrasensitive and Robust Assay of DNA Methyltransferase Activity
IF 5.7
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
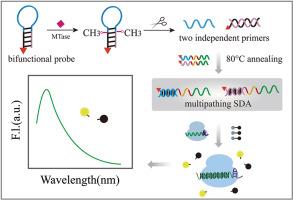
DNA methylation catalyzed by various DNA methyltransferases (DNA MTases) is one of the important epigenetic regulations in both eukaryotes and prokaryotes. Therefore, the detection of DNA MTase activity is a vital target and direction in the study of methylation-related diseases.Results
In this study, an ultrasensitive and robust strategy was developed for DNA MTase activity sensing based on bifunctional probe propelling multipath strand displacement amplification and CRISPR/Cas12a techniques. First, a bifunctional hairpin probe (bHpDNA) was designed instead of a conventional single-function probe. In the presence of DNA MTase, the bHpDNA was methylated and cleaved by a restriction endonuclease into two independent primers, both of which bind with the templates to trigger strand displacement amplification and produce the active DNA of CRISPR/Cas12a. Second, annealing-assisted binding instead of free diffusion adhesion was used to improve hybridization efficiency between the primers and templates. Finally, the CRISPR/Cas12a system was used to achieve fluorescence signal output to analyze DNA MTase activity. If targets were absent, there was no signal because no primers were released from the bHpDNA. To verify the reliability of the method, two key DNA MTases, Dam and M.SssI, were analyzed, and their limits of detection were 2.458 × 10−3 and 3.820 × 10−3 U/mL, respectively, which were lower than those of most reported fluorescence methods.Significance
This method was successfully used in the evaluation of DNA MTase inhibitors and the detection of DNA MTase activity in complex biological systems with good recoveries and relative standard deviation at low spiked concentrations (0.1–1 U/mL), which all indicate that this method is an ultrasensitive and robust strategy in DNA MTase activity assay and has great potential in biomedical and clinical detection.
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


