Subtype-specific transcription factors affect polyamine metabolism and the tumor microenvironment in breast cancer
Abstract
Background
Polyamines play important roles in cell growth and proliferation. Polyamine metabolism genes are dysregulated in various tumors. Some polyamine metabolism genes are regulated by transcription factors. However, the transcription factors that regulate polyamine metabolism genes have not been completely identified. Additionally, whether any of the transcriptional regulations depend on tumor heterogeneity and the tumor microenvironment has not been investigated.
Methods
We used bulk RNA-seq data to identify dysregulated polyamine metabolism genes and their transcription factors across breast cancer subtypes. Genes highly correlated with polyamine changes were obtained, and their subtype-specific expressions were checked in tumor microenvironment cells using single-cell RNA (scRNA)-seq data. Gene Ontology enrichment analysis was used to explore their molecular functions and biological processes, and survival analysis was used to examine the impact of these genes on therapeutic outcome.
Results
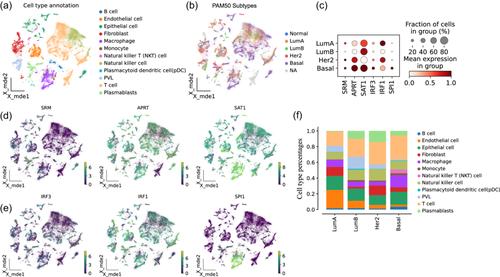
We first analyzed the dysregulation of polyamine synthesis, catabolism, and transport in four breast cancer subtypes. Genes such as AGMAT and CAV1 were dysregulated across all subtypes, while APRT, SAT1, and other genes were dysregulated in the more lethal subtypes. Among the dysregulated genes of polyamine metabolism, we focused on three genes (SRM, APRT, and SAT1) and identified their transcription factors (SPI1 and IRF1 correspond to SAT1, and IRF3 corresponds to SRM and APRT). With scRNA-seq data, we verified that these three transcription factors also regulated these three polyamine metabolism genes in the tumor microenvironment. Both bulk RNA-seq and scRNA-seq data indicated that these genes were specifically upregulated in high-risk breast cancer subtypes, such as the basal-like type. High expression of these genes corresponded to worse outcomes in the basal-like subtype under chemotherapy and radiation treatment.
Conclusion
Our work identified three subtype-specific transcription factors that regulate three polyamine metabolism genes in high-risk breast cancer subtypes and the tumor microenvironment. Our results deepen the understanding of the role of polyamine metabolism in breast cancer and may help the clinical therapy of advanced breast cancer subtypes.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


