The near-complete genome assembly of hexaploid wild oat reveals its genome evolution and divergence with cultivated oats
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
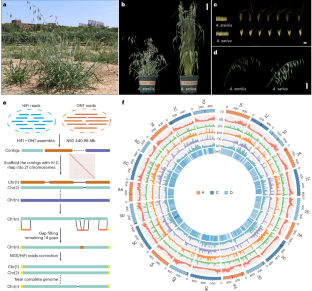
Avena sterilis, the ancestral species of cultivated oats, is a valuable genetic resource for oat improvement. Here we generated a near-complete 10.99 Gb A. sterilis genome and a high-quality 10.89 Gb cultivated oat genome. Genome evolution analysis revealed the centromeres dynamic and structural variations landscape associated with domestication between wild and cultivated oats. Population genetic analysis of 117 wild and cultivated oat accessions worldwide detected many candidate genes associated with important agronomic traits for oat domestication and improvement. Remarkably, a large fragment duplication from chromosomes 4A to 4D harbouring many agronomically important genes was detected during oat domestication and was fixed in almost all cultivated oats from around the world. The genes in the duplication region from 4A showed significantly higher expression levels and lower methylation levels than the orthologous genes located on 4D in A. sterilis. This study provides valuable resources for evolutionary and functional genomics and genetic improvement of oat. The near-complete genome of hexaploid wild oat, along with 117 global wild and cultivated accessions, reveals genome divergence between wild and cultivated oats and a large fragment duplication event from chromosomes 4A to 4D during oat domestication.

六倍体野生燕麦基因组的近完整组装揭示了其与栽培燕麦基因组的进化和分化
无菌燕麦是栽培燕麦的祖先种,是燕麦改良的宝贵遗传资源。在这里,我们获得了一个近乎完整的10.99 Gb的a . sterilis基因组和一个高质量的10.89 Gb的栽培燕麦基因组。基因组进化分析揭示了野生燕麦和栽培燕麦在驯化过程中着丝粒的动态和结构变化。对全球117个野生和栽培燕麦材料的群体遗传分析发现了许多与燕麦驯化和改良重要农艺性状相关的候选基因。值得注意的是,在燕麦驯化过程中,从染色体4A到4D中发现了一个包含许多重要农学基因的大片段重复,并且在世界上几乎所有栽培燕麦中都被固定下来。与位于4D的同源基因相比,4A重复区基因的表达水平显著提高,甲基化水平显著降低。该研究为燕麦进化和功能基因组学以及遗传改良提供了宝贵的资源。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


