Single-cell RNA sequencing of peripheral blood links cell-type-specific regulation of splicing to autoimmune and inflammatory diseases
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Alternative splicing contributes to complex traits, but whether this differs in trait-relevant cell types across diverse genetic ancestries is unclear. Here we describe cell-type-specific, sex-biased and ancestry-biased alternative splicing in ~1 M peripheral blood mononuclear cells from 474 healthy donors from the Asian Immune Diversity Atlas. We identify widespread sex-biased and ancestry-biased differential splicing, most of which is cell-type-specific. We identify 11,577 independent cis-splicing quantitative trait loci (sQTLs), 607 trans-sGenes and 107 dynamic sQTLs. Colocalization between cis-eQTLs and trans-sQTLs revealed a cell-type-specific regulatory relationship between HNRNPLL and PTPRC. We observed an enrichment of cis-sQTL effects in autoimmune and inflammatory disease heritability. Specifically, we functionally validated an Asian-specific sQTL disrupting the 5′ splice site of TCHP exon 4 that putatively modulates the risk of Graves’ disease in East Asian populations. Our work highlights the impact of ancestral diversity on splicing and provides a roadmap to dissect its role in complex diseases at single-cell resolution. This analysis of single-cell RNA sequencing data from peripheral blood mononuclear cells for 474 individuals of diverse Asian ancestries in the Asian Immune Diversity Atlas links cell-type-specific splicing variation with autoimmune and inflammatory disease risk.
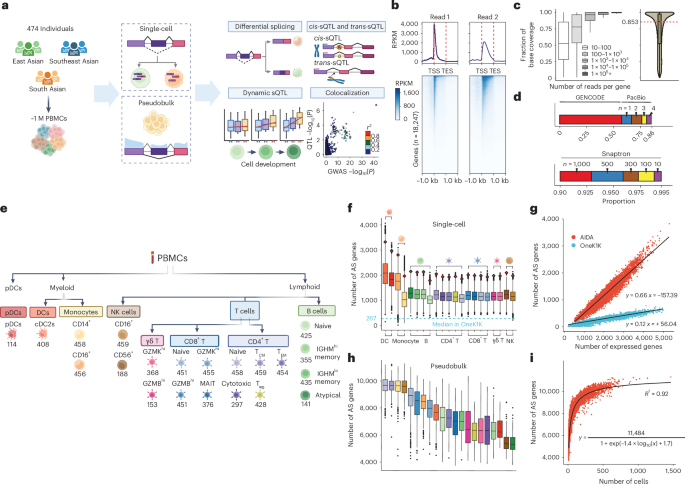

外周血单细胞RNA测序将细胞类型特异性剪接调节与自身免疫性和炎症性疾病联系起来
选择性剪接有助于形成复杂的性状,但在不同遗传祖先的性状相关细胞类型中是否存在差异尚不清楚。在这里,我们描述了来自亚洲免疫多样性图谱的474名健康供者的约1 M外周血单个核细胞的细胞类型特异性、性别偏倚和祖先偏倚的选择性剪接。我们发现广泛存在性别偏见和祖先偏见的差异剪接,其中大多数是细胞类型特异性的。共鉴定出11577个独立的顺式剪接数量性状位点(sqtl)、607个反式基因和107个动态sqtl。顺式- eqtl和反式- sqtl之间的共定位揭示了HNRNPLL和PTPRC之间的细胞类型特异性调控关系。我们观察到在自身免疫和炎症性疾病遗传力中,顺式- sqtl效应的富集。具体来说,我们从功能上验证了一个亚洲特异性的sQTL,该sQTL破坏了TCHP外显子4的5 '剪接位点,该位点被认为调节了东亚人群中Graves病的风险。我们的工作强调了祖先多样性对剪接的影响,并提供了在单细胞分辨率上解剖其在复杂疾病中的作用的路线图。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


