Machine Learning-Assisted, Dual-Channel CRISPR/Cas12a Biosensor-In-Microdroplet for Amplification-Free Nucleic Acid Detection for Food Authenticity Testing
IF 15.8
1区 材料科学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
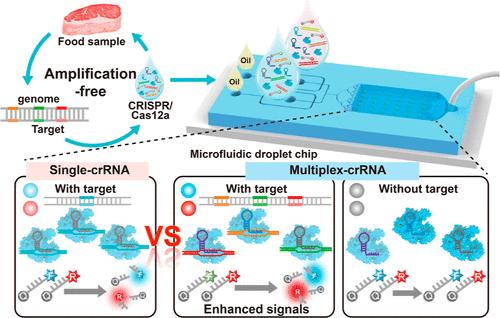
The development of novel detection technology for meat species authenticity is imperative. Here, we developed a machine learning-supported, dual-channel biosensor-in-microdroplet platform for meat species authenticity detection named CC-drop (CRISPR/Cas12a digital single-molecule microdroplet biosensor). This strategy allowed us to quickly identify and analyze animal-derived components in foods. This biosensor was enabled by CRISPR/Cas12a-based fluorescence lighting-up detection, and the nucleic acid signals can be recognized by a Cas12a–crRNA binary complex to trigger the trans-cleavage of any by-stander reporter single-stranded (ss) DNA, in which nucleic acid signals can be converted and amplified to fluorescent readouts. The ultralocalized microdroplet reactor was constructed by reducing the reaction volume from up to picoliter to accommodate the aforementioned reaction to further enhance the sensitivity to even render an amplification-free nucleic acid detection. Moreover, we established a smartphone App coupled with a random forest machine learning model based on parameters such as area, fluorescent intensity, and counting number to ensure the accuracy of image recording and processing. The sample-to-result time was within 80 min. Importantly, the proposed biosensor was able to accurately detect the ND1 (pork-specific) and IL-2 (duck-specific) genes in deep processed meat-derived foods that usually had truncated DNA, and the results were more robust and practical than conventional real-time polymerase chain reaction after a side-by-side comparison. All in all, the proposed biosensor can be expected to be used for rapid food authenticity and other nucleic acid detections in the future.
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Nano
工程技术-材料科学:综合
CiteScore
26.00
自引率
4.10%
发文量
1627
审稿时长
1.7 months
期刊介绍:
ACS Nano, published monthly, serves as an international forum for comprehensive articles on nanoscience and nanotechnology research at the intersections of chemistry, biology, materials science, physics, and engineering. The journal fosters communication among scientists in these communities, facilitating collaboration, new research opportunities, and advancements through discoveries. ACS Nano covers synthesis, assembly, characterization, theory, and simulation of nanostructures, nanobiotechnology, nanofabrication, methods and tools for nanoscience and nanotechnology, and self- and directed-assembly. Alongside original research articles, it offers thorough reviews, perspectives on cutting-edge research, and discussions envisioning the future of nanoscience and nanotechnology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


