Substrate translocation and inhibition in human dicarboxylate transporter NaDC3
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
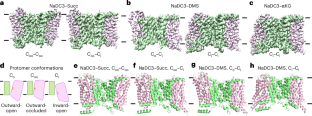
The human high-affinity sodium–dicarboxylate cotransporter (NaDC3) imports various substrates into the cell as tricarboxylate acid cycle intermediates, lipid biosynthesis precursors and signaling molecules. Understanding the cellular signaling process and developing inhibitors require knowledge of the structural basis of the dicarboxylate specificity and inhibition mechanism of NaDC3. To this end, we determined the cryo-electron microscopy structures of NaDC3 in various dimers, revealing the protomer in three conformations: outward-open Co, outward-occluded Coo and inward-open Ci. A dicarboxylate is first bound and recognized in Co and how the substrate interacts with NaDC3 in Coo likely helps to further determine the substrate specificity. A phenylalanine from the scaffold domain interacts with the bound dicarboxylate in the Coo state and modulates the kinetic barrier to the transport domain movement. Structural comparison of an inhibitor-bound structure of NaDC3 to that of the sodium-dependent citrate transporter suggests ways for making an inhibitor that is specific for NaDC3. The authors show cryo-electron microscopy structures of the human high-affinity sodium–dicarboxylate cotransporter, responsible for dicarboxylate import into the cell, in complex with various substrates and in different states establish the basis of substrate recognition, differentiation and release, as well as regulation of transport domain movement.

人二羧酸转运体NaDC3的底物易位和抑制作用
人类高亲和力钠-二羧酸盐共转运体(NaDC3)作为三羧酸酸循环中间体、脂质生物合成前体和信号分子将各种底物导入细胞。了解细胞信号传导过程和开发抑制剂需要了解NaDC3二羧酸盐特异性的结构基础和抑制机制。为此,我们测定了NaDC3在不同二聚体中的低温电镜结构,揭示了三种构象:向外开放的Co,向外封闭的Coo和向内开放的Ci。二羧酸盐首先在Co中结合并被识别,底物如何与Coo中的NaDC3相互作用可能有助于进一步确定底物特异性。来自支架结构域的苯丙氨酸在Coo状态下与结合的二羧酸盐相互作用,并调节转运结构域运动的动力学屏障。将NaDC3的抑制剂结合结构与钠依赖性柠檬酸转运体的结构进行比较,提出了制造NaDC3特异性抑制剂的方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


