Elucidation and engineering of Sphingolipid biosynthesis pathway in Yarrowia lipolytica for enhanced production of human-type sphingoid bases and glucosylceramides
IF 6.8
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Sphingolipids are vital membrane components in in mammalian cells, plants, and various microbes. We aimed to explore and exploit the sphingolipid biosynthesis pathways in an oleaginous and dimorphic yeast Yarrowia lipolytica by constructing and characterizing mutant strains with specific gene deletions and integrating exogenous genes to enhance the production of long-chain bases (LCBs) and glucosylceramides (GlcCers). To block the fungal/plant-specific phytosphingosine (PHS) pathway, we deleted the SUR2 gene encoding a sphinganine C4-hydroxylase, resulting in a remarkably elevated secretory production of dihydrosphingosine (DHS) and sphingosine (So) without acetylation. The Y. lipolytica SUR2 deletion (Ylsur2Δ) strain displayed retarded growth, increased pseudohyphal formation and stress sensitivity, along with the altered profiles of inositolphosphate-containing ceramides, GlcCers, and sterols. The subsequent disruption of the SLD1 gene, encoding a fungal/plant-specific Δ8 sphingolipid desaturase, restored filamentous growth in the Ylsur2Δ strain to a yeast-type form and further increased the production of human-type GlcCers. Additional introduction of mouse alkaline ceramidase 1 (maCER1) into the Ylsur2Δsld1Δ double mutants considerably increased DHS and So production while decreasing GlcCers. The production yields of LCBs from the Ylsur2Δsld1Δ/maCER1 strain increased in proportion to the C/N ratio in the N-source optimized medium, leading to production of 1.4 g/L non-acetylated DHS at the 5 L fed-batch fermentation with glucose feeding. This study highlights the feasibility of using the engineered Y. lipolytica strains as a cell factory for valuable sphingolipid derivatives for pharmaceuticals, cosmeceuticals, and nutraceuticals.
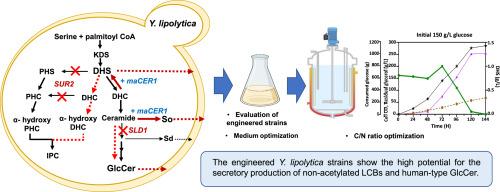
阐明并改造脂肪分解酵母中的鞘脂生物合成途径,以提高人型鞘氨醇碱和葡萄糖甘油酯的产量。
鞘脂是哺乳动物细胞、植物和各种微生物的重要膜成分。我们的目的是通过构建和鉴定特定基因缺失的突变株,并整合外源基因来提高长链碱和葡萄糖甘油酯的产量,从而探索和利用含油二形酵母亚罗酵母(Yarrowia lipolytica)的鞘脂生物合成途径。为了阻断真菌/植物特有的植物鞘氨醇(PHS)途径,我们删除了编码鞘氨醇 C4-羟化酶的 SUR2 基因,结果导致二氢鞘氨醇(DHS)和鞘氨醇(So)的分泌量显著增加,且没有乙酰化。脂溶性酵母菌 SUR2 基因缺失(Ylsur2Δ)菌株表现出生长迟缓、假茎形成增加和应激敏感性,以及含肌醇磷脂的神经酰胺、GlcCers 和固醇谱的改变。随后破坏了编码真菌/植物特异性Δ8鞘脂去饱和酶的 SLD1 基因,使 Ylsur2Δ 菌株的丝状生长恢复到酵母型,并进一步增加了人类型 GlcCers 的产量。在 Ylsur2Δsld1Δ 双突变体中引入小鼠碱性神经酰胺酶 1(maCER1)后,DHS 和 So 的产量显著增加,而 GlcCers 的产量则有所下降。Ylsur2Δsld1Δ/maCER1菌株的低氯苯产量与N源优化培养基中的C/N比成正比增加,导致在5升喂料批次发酵中葡萄糖喂养下的非乙酰化DHS产量达到1.4克/升。这项研究强调了将工程化的 Y. lipolytica 菌株作为细胞工厂生产有价值的鞘脂衍生物的可行性,这些衍生物可用于制药、药妆和营养保健品。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Metabolic engineering
工程技术-生物工程与应用微生物
CiteScore
15.60
自引率
6.00%
发文量
140
审稿时长
44 days
期刊介绍:
Metabolic Engineering (MBE) is a journal that focuses on publishing original research papers on the directed modulation of metabolic pathways for metabolite overproduction or the enhancement of cellular properties. It welcomes papers that describe the engineering of native pathways and the synthesis of heterologous pathways to convert microorganisms into microbial cell factories. The journal covers experimental, computational, and modeling approaches for understanding metabolic pathways and manipulating them through genetic, media, or environmental means. Effective exploration of metabolic pathways necessitates the use of molecular biology and biochemistry methods, as well as engineering techniques for modeling and data analysis. MBE serves as a platform for interdisciplinary research in fields such as biochemistry, molecular biology, applied microbiology, cellular physiology, cellular nutrition in health and disease, and biochemical engineering. The journal publishes various types of papers, including original research papers and review papers. It is indexed and abstracted in databases such as Scopus, Embase, EMBiology, Current Contents - Life Sciences and Clinical Medicine, Science Citation Index, PubMed/Medline, CAS and Biotechnology Citation Index.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


