Deep mutation, insertion and deletion scanning across the Enterovirus A proteome reveals constraints shaping viral evolution
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
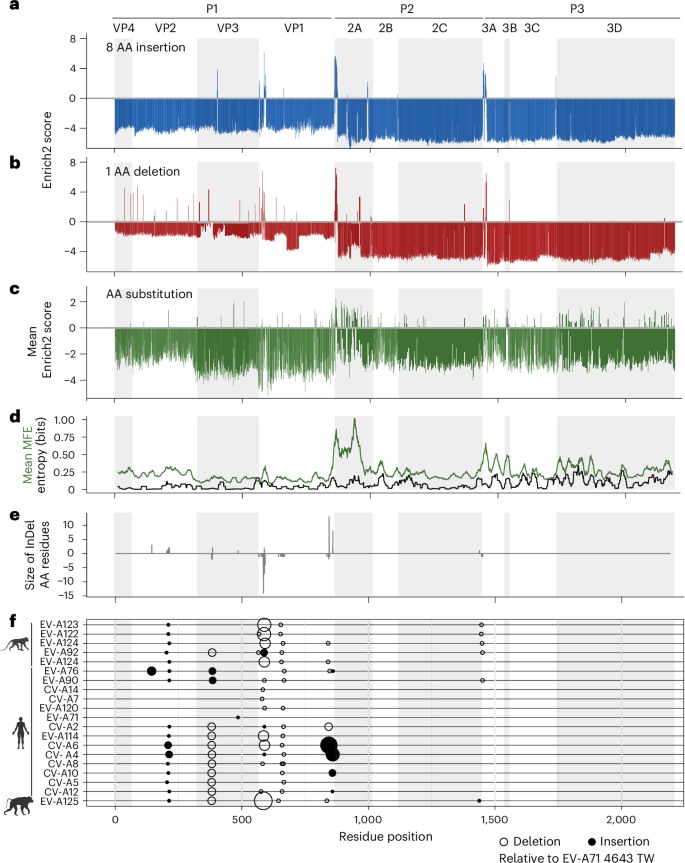
Insertions and deletions (InDels) are essential to protein evolution. In RNA viruses, InDels contribute to the emergence of viruses with new phenotypes, including altered host engagement and tropism. However, the tolerance of viral proteins for InDels has not been extensively studied. Here, we conduct deep mutational scanning to map and quantify the mutational tolerance of a complete viral proteome to insertion, deletion and substitution. We engineered approximately 45,000 insertions, 6,000 deletions and 41,000 amino acid substitutions across the nearly 2,200 coding positions of the Enterovirus A71 proteome, quantifying their effects on viral fitness by population sequencing. The vast majority of InDels are lethal to the virus, tolerated at only a few hotspots. Some of these hotspots overlap with sites of host recognition and immune engagement, suggesting tolerance at these sites reflects the important role InDels have played in the past phenotypic diversification of Enterovirus A. Deep mutational scanning of Enterovirus A71 maps and quantifies the impact of genomic insertions, deletions and substitutions on virus fitness and highlights the role of insertions and deletions in the diversification of RNA viruses.

对 A 型肠病毒蛋白质组进行深度突变、插入和缺失扫描,揭示影响病毒进化的制约因素
插入和缺失(InDels)对蛋白质进化至关重要。在 RNA 病毒中,InDels 有助于病毒产生新的表型,包括改变宿主参与和滋养性。然而,病毒蛋白质对 InDels 的耐受性尚未得到广泛研究。在这里,我们进行了深度突变扫描,以绘制和量化完整病毒蛋白质组对插入、缺失和置换的突变耐受性。我们在肠病毒 A71 蛋白组的近 2,200 个编码位置上设计了约 45,000 个插入位、6,000 个缺失位和 41,000 个氨基酸替换位,并通过群体测序量化了它们对病毒适应性的影响。绝大多数 InDels 对病毒都是致命的,只有少数几个热点可以容忍。其中一些热点与宿主识别和免疫参与位点重叠,表明这些位点的耐受性反映了 InDels 在肠病毒 A 过去的表型多样化过程中发挥的重要作用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


