Evolution and function of chromatin domains across the tree of life
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
The genome of all organisms is spatially organized to function efficiently. The advent of genome-wide chromatin conformation capture (Hi-C) methods has revolutionized our ability to probe the three-dimensional (3D) organization of genomes across diverse species. In this Review, we compare 3D chromatin folding from bacteria and archaea to that in mammals and plants, focusing on topology at the level of gene regulatory domains. In doing so, we consider systematic similarities and differences that hint at the origin and evolution of spatial chromatin folding and its relation to gene activity. We discuss the universality of spatial chromatin domains in all kingdoms, each encompassing one to several genes. We also highlight differences between organisms and suggest that similar features in Hi-C matrices do not necessarily reflect the same biological process or function. Furthermore, we discuss the evolution of domain boundaries and boundary-forming proteins, which indicates that structural maintenance of chromosome (SMC) proteins and the transcription machinery are the ancestral sculptors of the genome. Architectural proteins such as CTCF serve as clade-specific determinants of genome organization. Finally, studies in many non-model organisms show that, despite the ancient origin of 3D chromatin folding and its intricate link to gene activity, evolution tolerates substantial changes in genome organization. Szalay et al. discuss cross-kingdom similarities and differences in 3D chromatin folding in relation to gene regulation, including in bacteria, archaea, mammals and plants. This comparison reveals certain factors as ancestral sculptors of the genome, but also that evolution tolerates considerable variety in genome organization.
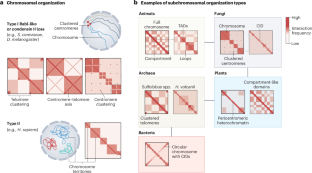

生命树上染色质结构域的进化与功能
所有生物的基因组都是按空间组织的,这样才能有效发挥作用。全基因组染色质构象捕获(Hi-C)方法的出现彻底改变了我们探究不同物种基因组三维组织的能力。在这篇综述中,我们将细菌和古细菌的三维染色质折叠与哺乳动物和植物的染色质折叠进行了比较,重点关注基因调控域水平的拓扑结构。在此过程中,我们考虑了系统性的相似之处和不同之处,这些相似之处和不同之处暗示了空间染色质折叠的起源和进化及其与基因活动的关系。我们讨论了空间染色质域在所有物种中的普遍性,每个染色质域包括一个到多个基因。我们还强调了生物体之间的差异,并指出Hi-C矩阵中的相似特征并不一定反映相同的生物过程或功能。此外,我们还讨论了结构域边界和边界形成蛋白的进化,这表明染色体结构维持(SMC)蛋白和转录机制是基因组的祖先雕刻者。CTCF等结构蛋白是基因组组织的特异性决定因素。最后,对许多非模式生物的研究表明,尽管三维染色质折叠起源古老,且与基因活动有着错综复杂的联系,但进化仍可容忍基因组组织发生重大变化。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


