A rapid, affordable, and reliable method for profiling microbiome biomarkers from fecal images
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Human and veterinary healthcare professionals are interested in utilizing the gut-microbiome as a target to diagnose, treat, and prevent (gastrointestinal) diseases. However, the current microbiome analysis techniques are expensive and time-consuming, and data interpretation requires the expertise of specialists. Therefore, we explored the development and application of artificial intelligence technology for rapid, affordable, and reliable microbiome profiling in rhesus macaques (Macaca mulatta). Tailor-made learning algorithms were created by integrating digital images of fecal samples with corresponding whole-genome sequenced microbial profiles. These algorithms were trained to identify alpha-diversity (Shannon index), key microbial markers, and fecal consistency from the digital images of fecal smears. A binary classification strategy was applied to distinguish between samples with high and low diversity and presence or absence of selected bacterial genera. Our results revealed a successful proof of concept for “high and low” prediction of diversity, fecal consistency, and “present or absent” for selected bacterial genera.
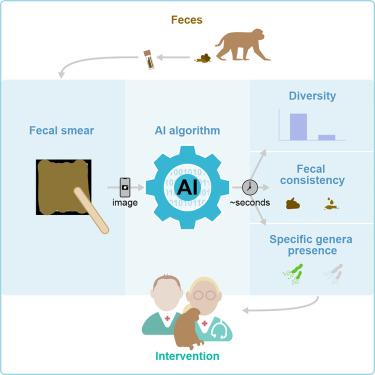
快速、经济、可靠的粪便图像微生物组生物标记分析方法
人类和兽医保健专业人员对利用肠道微生物组作为诊断、治疗和预防(胃肠道)疾病的目标很感兴趣。然而,目前的微生物组分析技术既昂贵又耗时,而且数据解读需要专家的专业知识。因此,我们探索了人工智能技术的开发和应用,以实现快速、经济、可靠的猕猴(Macaca mulatta)微生物组分析。通过整合粪便样本的数字图像和相应的全基因组测序微生物图谱,我们创建了量身定制的学习算法。对这些算法进行了训练,以便从粪便涂片的数字图像中识别阿尔法多样性(香农指数)、关键微生物标记物和粪便一致性。采用二元分类策略来区分样本的多样性高低和所选细菌属的存在与否。我们的研究结果表明,我们成功地验证了 "高低 "多样性预测、粪便一致性预测和选定细菌属 "存在或不存在 "预测的概念。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


