ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Gene expression is controlled by dynamic localization of thousands of regulatory proteins to precise genomic regions. Understanding this cell type-specific process has been a longstanding goal yet remains challenging because DNA–protein mapping methods generally study one protein at a time. Here, to address this, we developed chromatin immunoprecipitation done in parallel (ChIP-DIP) to generate genome-wide maps of hundreds of diverse regulatory proteins in a single experiment. ChIP-DIP produces highly accurate maps within large pools (>160 proteins) for all classes of DNA-associated proteins, including modified histones, chromatin regulators and transcription factors and across multiple conditions simultaneously. First, we used ChIP-DIP to measure temporal chromatin dynamics in primary dendritic cells following LPS stimulation. Next, we explored quantitative combinations of histone modifications that define distinct classes of regulatory elements and characterized their functional activity in human and mouse cell lines. Overall, ChIP-DIP generates context-specific protein localization maps at consortium scale within any molecular biology laboratory and experimental system. ChIP-DIP (ChIP done in parallel) is a highly multiplex assay for protein–DNA binding, scalable to hundreds of proteins including modified histones, chromatin regulators and transcription factors, offering a refined view of the cis-regulatory code.
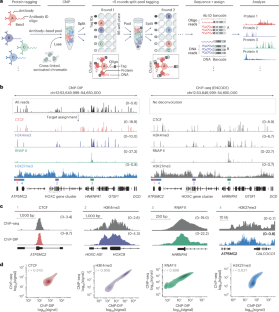

ChIP-DIP 可同时绘制出数百种蛋白质与 DNA 的结合图,并识别出各种基因调控元件
基因表达受控于数千种调控蛋白在精确基因组区域的动态定位。了解这一细胞类型特异性过程一直是我们的目标,但由于 DNA 蛋白图谱绘制方法通常一次只研究一种蛋白质,因此了解这一过程仍具有挑战性。为了解决这个问题,我们开发了染色质免疫共沉淀平行实验(ChIP-DIP),在一次实验中生成数百种不同调控蛋白的全基因组图谱。ChIP-DIP能在大量蛋白池(160个蛋白)中生成高度精确的图谱,涵盖所有类别的DNA相关蛋白,包括修饰组蛋白、染色质调控因子和转录因子,并能同时跨越多种条件。首先,我们使用 ChIP-DIP 测量了原代树突状细胞在受到 LPS 刺激后的染色质时间动态。接着,我们探索了组蛋白修饰的定量组合,这些组合定义了不同类别的调控元件,并描述了它们在人类和小鼠细胞系中的功能活性。总之,在任何分子生物学实验室和实验系统中,ChIP-DIP 都能在联合规模上生成特定背景的蛋白质定位图。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


