A multifaceted approach to understanding protein-buffer interactions in biopharmaceuticals
IF 4.4
2区 医学
Q1 PHARMACOLOGY & PHARMACY
European Journal of Pharmaceutics and Biopharmaceutics
Pub Date : 2024-11-19
DOI:10.1016/j.ejpb.2024.114582
引用次数: 0
Abstract
The excipient selection process plays a crucial role in biopharmaceutical formulation development to ensure the long-term stability of the drug product. Though there are numerous options approved by regulatory authorities, only a subset is commonly utilized. Previous research has proposed various stabilization mechanisms, including protein-excipient interactions. However, identifying these interactions remains challenging due to their weak and transient nature. In this study, we present a comprehensive approach to identify such interactions. Using the CPMG (Carr-Purcel-Meiboom-Gill) filter experiment we identified interactions of rituximab with certain buffers and amino acids, shedding light on its Fc fragment instability that manifested during the enzymatic cleavage of the antibody. Moreover, chemometric analyses of 2D NMR fingerprints revealed interactions of selected excipients with antibody fragments. Furthermore, molecular dynamics simulations revealed potential interacting hotspots without NMR spectra assignment. Our results highlight the importance of an orthogonal methods approach to uncovering these critical interactions, advancing our understanding of excipient stabilization mechanisms and rational formulation design in biopharmaceutics.
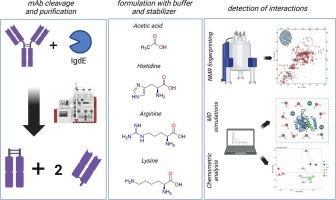
从多方面了解生物制药中蛋白质与缓冲剂的相互作用。
辅料选择过程在生物制药配方开发中起着至关重要的作用,可确保药物产品的长期稳定性。虽然监管机构批准的辅料有很多,但通常只使用其中的一部分。以往的研究提出了各种稳定机制,包括蛋白质与辅料之间的相互作用。然而,由于这些相互作用的微弱性和瞬时性,确定这些相互作用仍然具有挑战性。在本研究中,我们提出了一种识别此类相互作用的综合方法。利用 1HT2 CPMG(Carr-Purcel-Meiboom-Gill)过滤实验,我们确定了利妥昔单抗与某些缓冲液和氨基酸的相互作用,从而揭示了抗体在酶解过程中表现出的 Fc 片段不稳定性。此外,二维核磁共振指纹的化学计量分析表明了某些辅料与抗体片段的相互作用。此外,分子动力学模拟还揭示了潜在的相互作用热点,而无需核磁共振波谱赋值。我们的研究结果凸显了采用正交方法揭示这些关键相互作用的重要性,从而推动了我们对辅料稳定机制和生物制药领域合理配方设计的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.80
自引率
4.10%
发文量
211
审稿时长
36 days
期刊介绍:
The European Journal of Pharmaceutics and Biopharmaceutics provides a medium for the publication of novel, innovative and hypothesis-driven research from the areas of Pharmaceutics and Biopharmaceutics.
Topics covered include for example:
Design and development of drug delivery systems for pharmaceuticals and biopharmaceuticals (small molecules, proteins, nucleic acids)
Aspects of manufacturing process design
Biomedical aspects of drug product design
Strategies and formulations for controlled drug transport across biological barriers
Physicochemical aspects of drug product development
Novel excipients for drug product design
Drug delivery and controlled release systems for systemic and local applications
Nanomaterials for therapeutic and diagnostic purposes
Advanced therapy medicinal products
Medical devices supporting a distinct pharmacological effect.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


