Optimization of the activity and biodegradability of ionizable lipids for mRNA delivery via directed chemical evolution
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
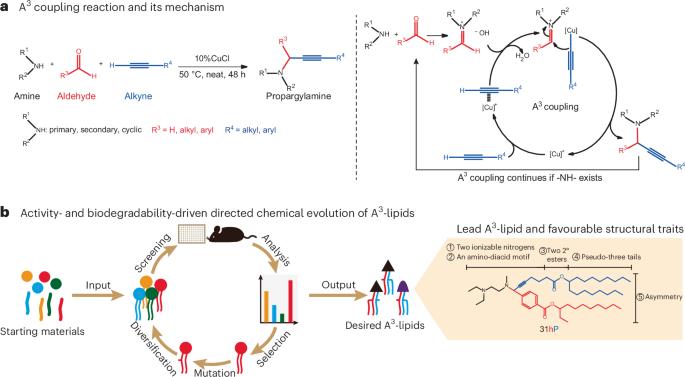
Ionizable lipids largely determine the biocompatibility of lipid nanoparticles (LNPs) and the efficacy for mRNA delivery. Rational design and combinatorial synthesis have led to the development of potent and biodegradable ionizable lipids, yet methodologies for the stepwise optimization of ionizable lipid structure are lacking. Here we show that iterative chemical derivatization and combinatorial chemistry, and in particular the amine–aldehyde–alkyne coupling reaction, can be leveraged to iteratively accelerate the structural optimization of propargylamine-based ionizable lipids (named A3-lipids) to improve their delivery activity and biodegradability. Through five cycles of such directed chemical evolution, we identified dozens of biodegradable and asymmetric A3-lipids with delivery activity comparable to or better than a benchmark ionizable lipid. We then derived structure−activity relationships for the headgroup, ester linkage and tail. Compared with standard ionizable lipids, the lead A3-lipid improved the hepatic delivery of an mRNA-based genome editor and the intramuscular delivery of an mRNA vaccine against SARS-CoV-2. Structural criteria for ionizable lipids discovered via directed chemical evolution may accelerate the development of LNPs for mRNA delivery. Directed chemical evolution can iteratively accelerate the structural optimization of ionizable lipids to improve their delivery activity and biodegradability for applications in lipid nanoparticle-mediated mRNA delivery.
通过定向化学进化优化可离子化脂质的活性和生物降解性,用于 mRNA 输送
可电离脂质在很大程度上决定了脂质纳米颗粒(LNPs)的生物相容性和 mRNA 递送的功效。合理的设计和组合合成已经开发出了强效和可生物降解的可离子化脂质,但目前还缺乏逐步优化可离子化脂质结构的方法。在这里,我们展示了迭代化学衍生和组合化学,特别是胺-醛-炔偶联反应,可用于加速丙炔胺基可离子化脂质(命名为 A3-脂质)的结构优化,以提高其递送活性和生物降解性。通过五个周期的定向化学进化,我们发现了数十种可生物降解的不对称 A3 脂,其递送活性与基准可电离脂相当或更好。然后,我们得出了头基、酯连接和尾部的结构-活性关系。与标准可离子化脂质相比,先导 A3 脂质改善了基于 mRNA 的基因组编辑器的肝脏递送和针对 SARS-CoV-2 的 mRNA 疫苗的肌肉递送。通过定向化学进化发现的可离子化脂质的结构标准可能会加速用于递送 mRNA 的 LNPs 的开发。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


