Using UK Biobank data to establish population-specific atlases from whole body MRI
IF 5.4
Q1 MEDICINE, RESEARCH & EXPERIMENTAL
引用次数: 0
Abstract
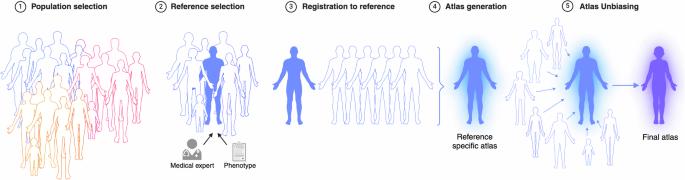
Reliable reference data in medical imaging is largely unavailable. Developing tools that allow for the comparison of individual patient data to reference data has a high potential to improve diagnostic imaging. Population atlases are a commonly used tool in medical imaging to facilitate this. Constructing such atlases becomes particularly challenging when working with highly heterogeneous datasets, such as whole-body images, which contain significant anatomical variations. In this work, we propose a pipeline for generating a standardised whole-body atlas for a highly heterogeneous population by partitioning the population into anatomically meaningful subgroups. Using magnetic resonance images from the UK Biobank dataset, we create six whole-body atlases representing a healthy population average. We furthermore unbias them, and this way obtain a realistic representation of the population. In addition to the anatomical atlases, we generate probabilistic atlases that capture the distributions of abdominal fat (visceral and subcutaneous) and five abdominal organs across the population (liver, spleen, pancreas, left and right kidneys). Our pipeline effectively generates high-quality, realistic whole-body atlases with clinical applicability. The probabilistic atlases show differences in fat distribution between subjects with medical conditions such as diabetes and cardiovascular diseases and healthy subjects in the atlas space. With this work, we make the constructed anatomical and label atlases publically available, with the expectation that they will support medical research involving whole-body MR images. Medical imaging requires examples of healthy images to be available for comparison with individual patient data. This comparison is important to detect changes that are indicative of disease and enable diagnosis. Population atlases consist of healthy images that can be used for comparisons. The images should match population characteristics as much as possible. However, building these atlases can be difficult, especially if the images used to compile the atlas show differences. In this study, we provide a method to create a standardised whole-body atlas using whole-body (neck to knee) magnetic resonance images. We produce a set of atlases that represent a healthy population. These atlases have been made publicly available and should assist medical researchers and improve healthcare outcomes for patients. Starck and Sideri-Lampretsa et al. propose a pipeline for generating whole-body atlases from a heterogeneous population by dividing it into anatomically meaningful subgroups. They demonstrate the use of these atlases for studying differences between healthy individuals and those with conditions such as diabetes or cardiovascular disease.
利用英国生物库数据,从全身核磁共振成像中建立特定人群图谱。
背景:医学成像中可靠的参考数据在很大程度上是不可用的。开发可将单个患者数据与参考数据进行比较的工具极有可能改进影像诊断。人群图谱是医学影像中常用的工具,可促进这一工作。在处理高度异构的数据集时,构建这样的图集尤其具有挑战性,例如包含显著解剖学差异的全身图像:在这项工作中,我们提出了一个管道,通过将人群划分为具有解剖意义的亚组,为高度异构的人群生成标准化的全身图集。利用英国生物库数据集的磁共振图像,我们创建了代表健康人群平均水平的六个全身图谱。此外,我们还对它们进行了不偏倚处理,从而获得了一个真实的人群代表。除了解剖图集外,我们还生成了概率图集,以捕捉整个人群的腹部脂肪(内脏和皮下脂肪)和五个腹部器官(肝脏、脾脏、胰腺、左肾和右肾)的分布情况:结果:我们的管道能有效生成高质量、逼真的全身图谱,具有临床应用价值。概率图集显示了患有糖尿病和心血管疾病等疾病的受试者与健康受试者在图集空间的脂肪分布差异:通过这项工作,我们公开了所构建的解剖和标签图谱,希望它们能为涉及全身磁共振图像的医学研究提供支持。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


