An integrated transcriptomic cell atlas of human neural organoids
IF 50.5
1区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
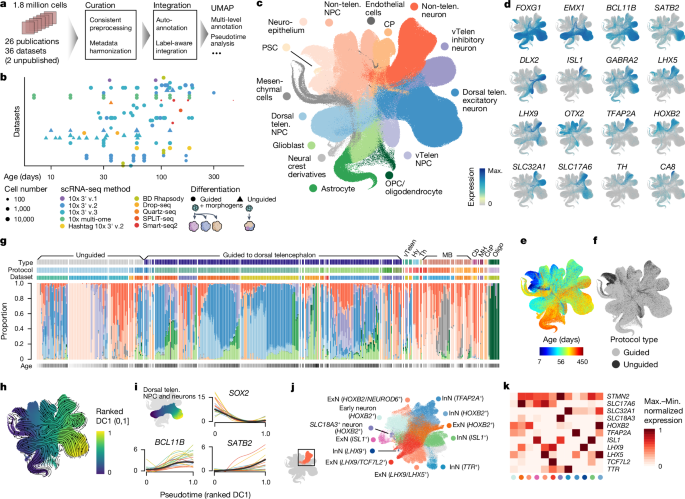
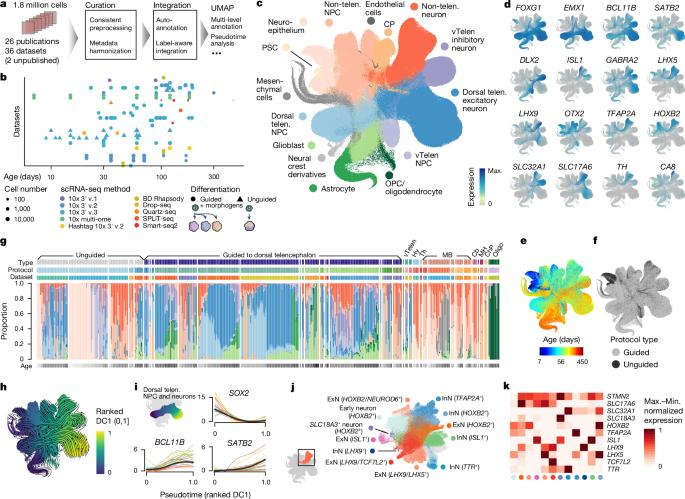
Human neural organoids, generated from pluripotent stem cells in vitro, are useful tools to study human brain development, evolution and disease. However, it is unclear which parts of the human brain are covered by existing protocols, and it has been difficult to quantitatively assess organoid variation and fidelity. Here we integrate 36 single-cell transcriptomic datasets spanning 26 protocols into one integrated human neural organoid cell atlas totalling more than 1.7 million cells1–26. Mapping to developing human brain references27–30 shows primary cell types and states that have been generated in vitro, and estimates transcriptomic similarity between primary and organoid counterparts across protocols. We provide a programmatic interface to browse the atlas and query new datasets, and showcase the power of the atlas to annotate organoid cell types and evaluate new organoid protocols. Finally, we show that the atlas can be used as a diverse control cohort to annotate and compare organoid models of neural disease, identifying genes and pathways that may underlie pathological mechanisms with the neural models. The human neural organoid cell atlas will be useful to assess organoid fidelity, characterize perturbed and diseased states and facilitate protocol development. A human neural organoid cell atlas integrating 36 single-cell transcriptomic datasets shows cell types and states and estimates transcriptomic similarity between primary and organoid counterparts, showing potential to assess organoid fidelity and facilitate protocol development.
人类神经器官组织的综合转录组细胞图谱
由体外多能干细胞生成的人类神经器官组织是研究人类大脑发育、进化和疾病的有用工具。然而,目前还不清楚现有方案涵盖了人脑的哪些部分,也很难定量评估类器官的变异和保真度。在这里,我们将跨越 26 个方案的 36 个单细胞转录组数据集整合到一个总计超过 170 万个细胞的人类神经类器官细胞图谱中1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26。与发育中人脑参考文献27,28,29,30 的映射显示了体外生成的原代细胞类型和状态,并估算了不同方案下原代细胞和类器官之间的转录组相似性。我们提供了浏览图谱和查询新数据集的程序界面,并展示了图谱在注释类器官细胞类型和评估新类器官方案方面的强大功能。最后,我们还展示了该图集可作为一个多样化的对照队列,用于注释和比较神经疾病的类器官模型,识别可能是神经模型病理机制基础的基因和通路。人类神经类器官细胞图谱将有助于评估类器官的保真度、描述紊乱和疾病状态以及促进方案开发。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature
综合性期刊-综合性期刊
CiteScore
90.00
自引率
1.20%
发文量
3652
审稿时长
3 months
期刊介绍:
Nature is a prestigious international journal that publishes peer-reviewed research in various scientific and technological fields. The selection of articles is based on criteria such as originality, importance, interdisciplinary relevance, timeliness, accessibility, elegance, and surprising conclusions. In addition to showcasing significant scientific advances, Nature delivers rapid, authoritative, insightful news, and interpretation of current and upcoming trends impacting science, scientists, and the broader public. The journal serves a dual purpose: firstly, to promptly share noteworthy scientific advances and foster discussions among scientists, and secondly, to ensure the swift dissemination of scientific results globally, emphasizing their significance for knowledge, culture, and daily life.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


