Ancestral Sequence Reconstruction Meets Machine Learning: Ene Reductase Thermostabilization Yields Enzymes with Improved Reactivity Profiles
IF 11.3
1区 化学
Q1 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
Ene reductases (EREDs) are enzymes that catalyze the asymmetric reduction of C═C bonds. EREDs are potentially useful in the large-scale synthesis of pharmaceutical compounds, but their application as biocatalysts is limited because they are often unstable under process conditions. Previous work addressed this limitation by identifying stabilized EREDs with ancestral sequence reconstruction (ASR), a bioinformatic method that predicts evolutionary ancestors based on a set of homologous sequences. In this work, we sought to apply ASR to design enzyme libraries and leverage machine learning to predict the most stable library variants. We generated an ERED library that targeted residues based on uncertainty in the ASR prediction. Screening data from a portion of the library were used to build a machine learning model that could accurately predict variants with improved thermostability. The most stabilized enzyme outperformed the wild-type and ancestral parent enzymes under process-like conditions with a panel of substrates. We envision that the combination of ASR and machine learning could be generally applied to other classes of enzymes, facilitating the development of high-quality industrial biocatalysts.
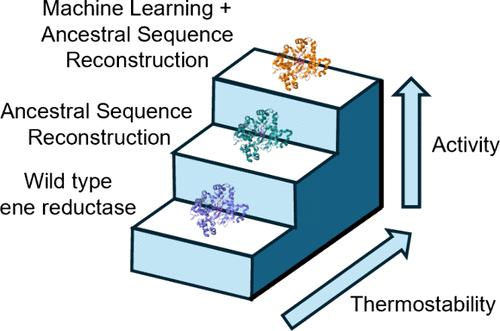
祖先序列重建与机器学习:炔还原酶热稳定化产生具有更佳反应谱的酶
炔还原酶(ERED)是催化 C═C 键不对称还原的酶。ERED在大规模合成药物化合物方面具有潜在的用途,但由于它们在加工条件下通常不稳定,因此其作为生物催化剂的应用受到了限制。以前的工作通过祖先序列重建(ASR)来识别稳定的 ERED,从而解决了这一局限性。ASR 是一种生物信息学方法,可根据一组同源序列预测进化祖先。在这项工作中,我们试图应用 ASR 设计酶库,并利用机器学习预测最稳定的库变体。我们根据 ASR 预测的不确定性生成了一个 ERED 库,以残基为目标。来自部分库的筛选数据被用来建立一个机器学习模型,该模型可以准确预测热稳定性更好的变体。在一组底物的类似过程条件下,最稳定的酶的表现优于野生型酶和祖先母酶。我们设想 ASR 与机器学习的结合可普遍应用于其他类别的酶,从而促进高质量工业生物催化剂的开发。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Catalysis
CHEMISTRY, PHYSICAL-
CiteScore
20.80
自引率
6.20%
发文量
1253
审稿时长
1.5 months
期刊介绍:
ACS Catalysis is an esteemed journal that publishes original research in the fields of heterogeneous catalysis, molecular catalysis, and biocatalysis. It offers broad coverage across diverse areas such as life sciences, organometallics and synthesis, photochemistry and electrochemistry, drug discovery and synthesis, materials science, environmental protection, polymer discovery and synthesis, and energy and fuels.
The scope of the journal is to showcase innovative work in various aspects of catalysis. This includes new reactions and novel synthetic approaches utilizing known catalysts, the discovery or modification of new catalysts, elucidation of catalytic mechanisms through cutting-edge investigations, practical enhancements of existing processes, as well as conceptual advances in the field. Contributions to ACS Catalysis can encompass both experimental and theoretical research focused on catalytic molecules, macromolecules, and materials that exhibit catalytic turnover.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


