Multiplex Digital PCR for the simultaneous quantification of a miRNA panel
IF 5.7
2区 化学
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
Background
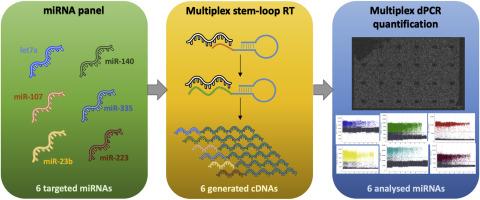
microRNAs (miRNAs) are small non-coding RNAs regulating gene expression. They have attracted significant interest as biomarkers for early diagnosis, prediction and monitoring of treatment response in many diseases. As individual miRNAs often lack the required sensitivity and specificity, miRNA signatures are developed for clinical applications. Digital PCR (dPCR) is a sensitive fluorescent-based quantification method, that can be used to detect the expression of miRNAs in patient samples. Our study presents the first proof-of-concept of a multiplexed dPCR assay for the simultaneous analysis and quantification of multiple miRNAs.Results
After reverse transcription (RT) using a pool of miRNA-specific stem-loop primers, dPCR was performed with a universal reverse primer and miRNA-specific forward primers along with fluorescently-labelled hydrolysis probes. Multiple experimental parameters were evaluated and strategies for modulating the observed signals were devised. The optimised assay was applied to the analysis of miRNAs from cell lines and biological samples. Although absolute quantification was lost, due to the reverse transcription step, quantification was linear for the dilution series and results were highly reproducible for independent dPCR and RT reactions. Our results confirmed the high sensitivity of dPCR for patient samples.Conclusions
We demonstrate the feasibility and reliability of multiplexed detection and quantification of miRNAs by dPCR that can be applied in a clinical setting to evaluate miRNA signatures.
用于同时定量 miRNA 面板的多重数字 PCR
背景微小核糖核酸(miRNA)是调节基因表达的小型非编码核糖核酸。作为许多疾病的早期诊断、预测和治疗反应监测的生物标志物,它们引起了人们的极大兴趣。由于单个 miRNA 通常缺乏所需的灵敏度和特异性,因此开发了用于临床应用的 miRNA 标志。数字 PCR(dPCR)是一种基于荧光的灵敏定量方法,可用于检测患者样本中 miRNA 的表达。结果在使用一组 miRNA 特异性茎环引物进行反转录(RT)后,使用通用反向引物和 miRNA 特异性正向引物以及荧光标记的水解探针进行了 dPCR。对多个实验参数进行了评估,并设计了调节观察到的信号的策略。优化后的检测方法被用于分析细胞系和生物样本中的 miRNA。虽然由于反转录步骤而失去了绝对定量,但稀释系列的定量是线性的,独立的 dPCR 和 RT 反应的结果具有很高的重现性。我们的结果证实了 dPCR 对患者样本的高灵敏度。结论我们证明了通过 dPCR 对 miRNA 进行多重检测和定量的可行性和可靠性,这种方法可用于临床环境中评估 miRNA 特征。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Analytica Chimica Acta
化学-分析化学
CiteScore
10.40
自引率
6.50%
发文量
1081
审稿时长
38 days
期刊介绍:
Analytica Chimica Acta has an open access mirror journal Analytica Chimica Acta: X, sharing the same aims and scope, editorial team, submission system and rigorous peer review.
Analytica Chimica Acta provides a forum for the rapid publication of original research, and critical, comprehensive reviews dealing with all aspects of fundamental and applied modern analytical chemistry. The journal welcomes the submission of research papers which report studies concerning the development of new and significant analytical methodologies. In determining the suitability of submitted articles for publication, particular scrutiny will be placed on the degree of novelty and impact of the research and the extent to which it adds to the existing body of knowledge in analytical chemistry.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


