Light-controlled protein imprinted nanospheres with variable recognition specificity
IF 8.9
1区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
This work develops a protein imprinted nanosphere with varied recognition specificity for bovine serum albumin (BSA) and lysozyme (Lyz) under different UV light through a gradient dual crosslinked imprinting strategy (i.e., covalent crosslinking and dynamic reversible crosslinking). The imprinting cavities are initially constructed using irreversible covalent crosslinking to specifically recognize BSA, and then the coumarin residues in the imprinting cavities are crosslinked under 365 nm UV light to further imprint Lyz, because Lyz has smaller size than BSA. Since the photo-crosslinking of coumarin is a reversible reaction, the imprinting cavities of Lyz can be de-crosslinked under 254 nm UV light and restore the imprinting cavities of BSA. Moreover, the N-isopropyl acrylamide (NIPAM) and pyrrolidine residues copolymerized in the polymeric surface of the nanospheres are temperature- and pH-responsive respectively. Therefore, the protein rebinding and release behaviors of the nanospheres are controlled by external temperature and pH. As a result, the materials can selectively separate BSA from real bovine whole blood and Lyz from egg white under different UV light. This study may provide a new strategy for construction of protein imprinted materials with tunable specificity for different proteins.
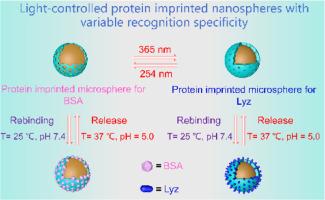
具有可变识别特异性的光控蛋白印迹纳米球
这项研究通过梯度双交联印迹策略(即共价交联和动态可逆交联),开发了一种在不同紫外光下对牛血清白蛋白(BSA)和溶菌酶(Lyz)具有不同识别特异性的蛋白质印迹纳米球。首先利用不可逆共价交联构建印记腔,以特异性识别 BSA,然后在 365 纳米紫外光下交联印记腔中的香豆素残基,进一步印记 Lyz,因为 Lyz 的尺寸比 BSA 小。由于香豆素的光交联是一种可逆反应,因此在 254 纳米紫外光下,Lyz 的印迹空腔可以去交联,恢复 BSA 的印迹空腔。此外,纳米球聚合物表面共聚的 N-异丙基丙烯酰胺(NIPAM)和吡咯烷残基分别具有温度和 pH 响应性。因此,纳米球的蛋白质再结合和释放行为受外部温度和 pH 值的控制。因此,在不同的紫外光下,该材料可选择性地从真正的牛全血中分离出 BSA,从蛋清中分离出 Lyz。这项研究为构建对不同蛋白质具有可调特异性的蛋白质印迹材料提供了一种新策略。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Chinese Chemical Letters
化学-化学综合
CiteScore
14.10
自引率
15.40%
发文量
8969
审稿时长
1.6 months
期刊介绍:
Chinese Chemical Letters (CCL) (ISSN 1001-8417) was founded in July 1990. The journal publishes preliminary accounts in the whole field of chemistry, including inorganic chemistry, organic chemistry, analytical chemistry, physical chemistry, polymer chemistry, applied chemistry, etc.Chinese Chemical Letters does not accept articles previously published or scheduled to be published. To verify originality, your article may be checked by the originality detection service CrossCheck.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


