Stronger binding affinities of gp120/CD4 in Catarrhini provide insights into HIV/host interactions
IF 2.5
3区 医学
Q1 Medicine
引用次数: 0
Abstract
Human immunodeficiency virus-1 (HIV-1) exploits the viral gp120 protein and host CD4/CCR5 receptors for the pandemic infection to humans. The host co-receptors of not only humans but also several primates and HIV-model mice can interact with the HIV receptor. However, the molecular mechanisms of these interactions remain unclear. Using Shaik et al. (2019)'s gp120/CD4/CCR5 structure of HIV-1B and human, here, we investigate the molecular dynamics between HIV sub-lineages (B, C, N, and O) and potential hosts in Euarchontoglires (primates and rodents). Although both host genes show similar protein structures conserved in all animals, CD4 gene demonstrates significantly stronger binding affinities in Catarrhini (apes and Old-World monkeys). Its known candidate residues interacted with gp120 fail to explain these affinity variations. Therefore, we identified novel candidate sites under positive selection on the Catarrhini lineage. Among four positively selected sites, residue R58 in humans is located within an antigen-antibody binding domain, exhibiting apomorphic amino acid substitutions as Arginine (R) in Catarrhini, which are mutually exclusive to the other animals where Lysine (K) is prevalent. Applying for artificial mutation test, we validated that K to R substitutions can lead stronger binding affinities of Catarrhini. Ecologically, these dynamics may relate to shared equatorial habitats in Africa and Asia. Our findings suggest a new candidate site R58 driven by the lineage-specific evolution as a molecular foundation on HIV infection.
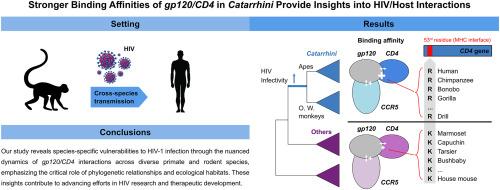
猫科动物体内 gp120/CD4 更强的结合亲和力有助于深入了解艾滋病毒与宿主的相互作用
人类免疫缺陷病毒-1(HIV-1)利用病毒 gp120 蛋白和宿主 CD4/CCR5 受体对人类进行大流行感染。不仅人类,一些灵长类动物和艾滋病毒模型小鼠的宿主共受体也能与艾滋病毒受体相互作用。然而,这些相互作用的分子机制仍不清楚。利用 Shaik 等人(2019 年)的 HIV-1B 和人类 gp120/CD4/CCR5 结构,我们在此研究了 HIV 亚系(B、C、N 和 O)与欧亚动物(灵长类和啮齿类)潜在宿主之间的分子动态。虽然两种宿主基因都显示出在所有动物中都保留的类似蛋白质结构,但 CD4 基因在鼬科动物(猿和旧世界猴)中显示出明显更强的结合亲和力。其与 gp120 相互作用的已知候选残基无法解释这些亲和力变化。因此,我们在猫科动物中发现了正向选择的新候选位点。在四个正选择位点中,人类的残基 R58 位于抗原-抗体结合域内,在猫科动物中表现为精氨酸(R)的非形态氨基酸取代,这与其他动物中赖氨酸(K)的普遍存在相互排斥。通过人工突变测试,我们验证了 K 到 R 的置换可导致 Catarrhini 更强的结合亲和力。从生态学角度看,这些动态可能与非洲和亚洲共同的赤道栖息地有关。我们的研究结果表明,一个新的候选位点 R58 是由血统特异性进化驱动的,是 HIV 感染的分子基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Infectious Disease Modelling
Mathematics-Applied Mathematics
CiteScore
17.00
自引率
3.40%
发文量
73
审稿时长
17 weeks
期刊介绍:
Infectious Disease Modelling is an open access journal that undergoes peer-review. Its main objective is to facilitate research that combines mathematical modelling, retrieval and analysis of infection disease data, and public health decision support. The journal actively encourages original research that improves this interface, as well as review articles that highlight innovative methodologies relevant to data collection, informatics, and policy making in the field of public health.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


