Deep learning-based hyperspectral image correction and unmixing for brain tumor surgery
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
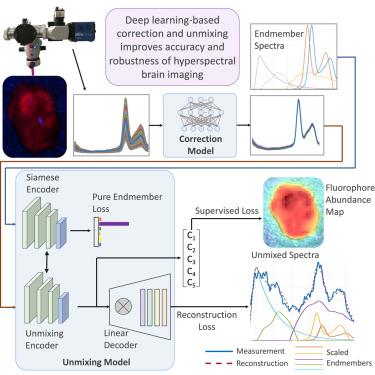
Hyperspectral imaging for fluorescence-guided brain tumor resection improves visualization of tissue differences, which can ameliorate patient outcomes. However, current methods do not effectively correct for heterogeneous optical and geometric tissue properties, leading to less accurate results. We propose two deep learning models for correction and unmixing that can capture these effects. While one is trained with protoporphyrin IX (PpIX) concentration labels, the other is semi-supervised. The models were evaluated on phantom and pig brain data with known PpIX concentration; the supervised and semi-supervised models achieved Pearson correlation coefficients (phantom, pig brain) between known and computed PpIX concentrations of (0.997, 0.990) and (0.98, 0.91), respectively. The classical approach achieved (0.93, 0.82). The semi-supervised approach also generalizes better to human data, achieving a 36% lower false-positive rate for PpIX detection and giving qualitatively more realistic results than existing methods. These results show promise for using deep learning to improve hyperspectral fluorescence-guided neurosurgery.
基于深度学习的脑肿瘤手术高光谱图像校正和非混合技术
用于荧光引导脑肿瘤切除术的高光谱成像技术可改善组织差异的可视化,从而改善患者的预后。然而,目前的方法不能有效校正异质光学和几何组织特性,导致结果不够准确。我们提出了两种用于校正和解除混合的深度学习模型,可以捕捉到这些影响。其中一个是用原卟啉 IX(PpIX)浓度标签训练的,另一个是半监督的。这些模型在已知 PpIX 浓度的模型和猪脑数据上进行了评估;监督和半监督模型在已知和计算的 PpIX 浓度之间的皮尔逊相关系数(模型、猪脑)分别为(0.997,0.990)和(0.98,0.91)。经典方法达到了(0.93, 0.82)。半监督方法对人类数据的泛化效果也更好,PpIX 检测的假阳性率降低了 36%,与现有方法相比,半监督方法得到了更真实的结果。这些结果表明,利用深度学习改进高光谱荧光引导神经外科手术大有可为。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


