Exploring structured molecular landscape from single-cell multi-omics data by an explainable multimodal model
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
There is an urgent need to understand the molecular landscape beyond the conventional cellular landscape, maximizing the translational use and generalized interpretation of state-of-the-art single-cell genomic techniques in biological studies. We introduced a multimodal explainable artificial intelligence (xAI) model Vec3D to identify a joint definition of cellular states and their distribution in a quantified graphic organization as structured molecular landscape (SML). First, Vec3D substantially improves the accuracy and efficiency of multimodal data analysis. Further, an SML was learned on CITE-seq data of human peripheral blood mononuclear cells (PBMCs), simultaneously revealing the predictive multi-label cell state and corresponding joint cell state markers with complementary effects from genes and proteins. Third, Vec3D demonstrated that the spatial-temporal SML efficiently characterizes molecular dynamics of cell lineages during human lung development. Collectively, Vec3D will be a broadly applicable computational method in the principle of “AI-for-biology”, providing a unified framework for understanding cellular homeostasis and imbalance through SML dynamics.
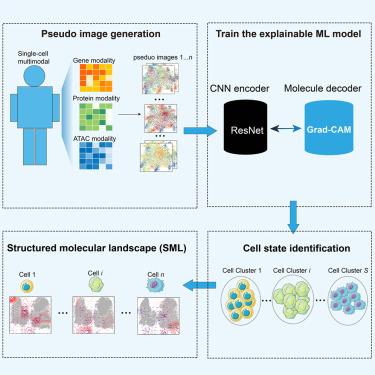
通过可解释的多模态模型从单细胞多组学数据中探索结构化分子景观
目前迫切需要了解传统细胞景观之外的分子景观,从而最大限度地在生物学研究中转化使用和通用解释最先进的单细胞基因组技术。我们引入了多模态可解释人工智能(xAI)模型 Vec3D,以确定细胞状态的联合定义及其在量化图形组织中的分布,即结构化分子景观(SML)。首先,Vec3D 大大提高了多模态数据分析的准确性和效率。其次,Vec3D 对人类外周血单核细胞(PBMCs)的 CITE-seq 数据进行了 SML 学习,同时揭示了预测性多标签细胞状态和相应的联合细胞状态标记,以及基因和蛋白质的互补效应。第三,Vec3D 证明了时空 SML 能有效描述人类肺部发育过程中细胞系的分子动力学特征。总之,Vec3D 将成为 "AI-for-biology "原则下一种广泛适用的计算方法,为通过 SML 动力学理解细胞平衡和失衡提供一个统一的框架。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


