Revealing taxonomy, activity, and substrate assimilation in mixed bacterial communities by GroEL-proteotyping-based stable isotope probing
IF 4.6
2区 综合性期刊
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
Protein-based stable isotope probing (protein-SIP) can link microbial taxa to substrate assimilation. Traditionally, protein-SIP requires a sample-specific metagenome-derived database for samples with unknown composition. Here, we describe GroEL-prototyping-based stable isotope probing (GroEL-SIP), that uses GroEL as a taxonomic marker protein to identify bacterial taxa (GroEL-proteotyping) coupled to SIP directly linking identified taxa to substrate consumption. GroEL-SIP’s main advantages are that (1) it can be performed with a sample-independent database and (2) sample complexity can be reduced by enriching GroEL proteins, increasing sensitivity and reducing instrument time. We applied GroEL-SIP to pure cultures, synthetic bicultures, and a human gut model using 2H-, 18O-, and 13C-labeled substrates. While 2H and 18O allowed assessing general activity, 13C enabled differentiation of substrate source and utilized metabolic pathways. GroEL-SIP offers fast and straightforward protein-SIP analyses of highly abundant families in mixed bacterial communities, but further work is needed to improve sensitivity, resolution, and database coverage.
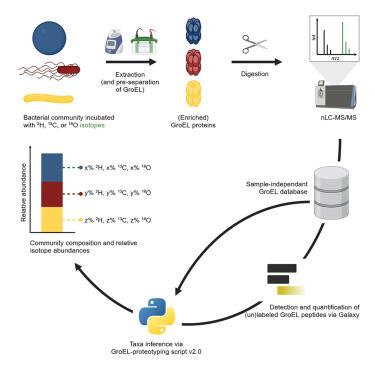
通过基于 GroEL 蛋白分型的稳定同位素探测揭示混合细菌群落的分类、活性和底物同化作用
基于蛋白质的稳定同位素探测(protein-SIP)可以将微生物类群与底物同化联系起来。传统上,蛋白质-SIP 需要一个特定于样本的元基因组数据库,用于未知成分的样本。在这里,我们介绍了基于GroEL-prototyping的稳定同位素探针(GroEL-SIP),它使用GroEL作为分类标记蛋白来识别细菌类群(GroEL-proteotyping),并结合SIP直接将已识别的类群与底物消耗联系起来。GroEL-SIP 的主要优点是:(1) 可使用与样品无关的数据库;(2) 可通过富集 GroEL 蛋白降低样品的复杂性,提高灵敏度并缩短仪器时间。我们使用 2H、18O 和 13C 标记的底物将 GroEL-SIP 应用于纯培养物、合成双培养物和人体肠道模型。2H 和 18O 可以评估一般活性,而 13C 则可以区分底物来源并利用代谢途径。GroEL-SIP为混合细菌群落中的高丰度家族提供了快速、直接的蛋白质-SIP分析,但还需要进一步的工作来提高灵敏度、分辨率和数据库覆盖率。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

iScience
Multidisciplinary-Multidisciplinary
CiteScore
7.20
自引率
1.70%
发文量
1972
审稿时长
6 weeks
期刊介绍:
Science has many big remaining questions. To address them, we will need to work collaboratively and across disciplines. The goal of iScience is to help fuel that type of interdisciplinary thinking. iScience is a new open-access journal from Cell Press that provides a platform for original research in the life, physical, and earth sciences. The primary criterion for publication in iScience is a significant contribution to a relevant field combined with robust results and underlying methodology. The advances appearing in iScience include both fundamental and applied investigations across this interdisciplinary range of topic areas. To support transparency in scientific investigation, we are happy to consider replication studies and papers that describe negative results.
We know you want your work to be published quickly and to be widely visible within your community and beyond. With the strong international reputation of Cell Press behind it, publication in iScience will help your work garner the attention and recognition it merits. Like all Cell Press journals, iScience prioritizes rapid publication. Our editorial team pays special attention to high-quality author service and to efficient, clear-cut decisions based on the information available within the manuscript. iScience taps into the expertise across Cell Press journals and selected partners to inform our editorial decisions and help publish your science in a timely and seamless way.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


