Quantitative analysis of dried serum FTIR spectra based on correlation Analysis-Interval random Frog-Partial least squares
IF 4.3
2区 化学
Q1 SPECTROSCOPY
Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy
Pub Date : 2024-11-10
DOI:10.1016/j.saa.2024.125427
引用次数: 0
Abstract
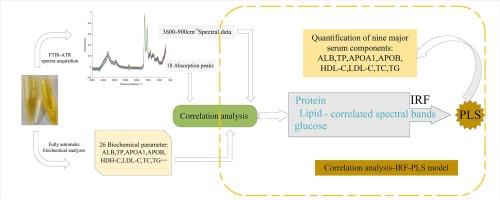
Serum biochemical markers are widely used in clinical practice but often require expensive, specific reagents, complex instruments, and prolonged result waiting times. Infrared spectroscopy offers multiple advantages for serum analysis, such as reagent-free testing and the ability to quickly and directly measure multiple parameters simultaneously. This study collected serum samples from 66 healthy subjects to explore the relationship between dried serum infrared spectra and biochemical parameters, and to investigate the feasibility of simultaneously quantifying nine major serum components using dried serum infrared spectra. Initially, correlation analysis was conducted between spectral data and biochemical parameters, and the correlation spectral bands of glucose, protein and lipid were determined according to the correlation results. Subsequently, the interval random frog (IRF) algorithm was utilized to select the optimal characteristic wavenumbers of the correlated spectral bands, extracting the most informative spectral variables and constructing partial least squares (PLS) quantitative models. This method successfully achieved rapid and accurate quantification of nine major components in serum, including glucose, total protein, albumin, apolipoprotein A1, apolipoprotein B, total cholesterol, triglycerides, low-density lipoprotein cholesterol, and high-density lipoprotein cholesterol. The experimental results showed that the correlation coefficient (Rp) range in the test set was 0.8892–0.9941. Among them, the quantification of total cholesterol yielded the highest Rp, corresponding to a root mean square error (RMSEP) of 7.2425 mg/dL in the test set, while the quantification of glucose yielded the lowest Rp, with an associated RMSEP of 2.3683 mg/dL. The Correlation Analysis (CA)-IRF-PLS method developed in this study outperformed the conventional PLS method, the direct use of the successive projection algorithm (SPA)-PLS quantitative method and other reported quantitative techniques, providing a novel approach for the real-time determination of clinical parameters in serum.
基于相关分析-间歇随机蛙-部分最小二乘法的干血清傅立叶变换红外光谱定量分析
血清生化指标被广泛应用于临床实践中,但通常需要昂贵的特异性试剂、复杂的仪器和较长的结果等待时间。红外光谱法在血清分析方面具有多种优势,如无需试剂检测,能够快速、直接地同时测量多个参数。本研究采集了 66 名健康受试者的血清样本,探讨了干血清红外光谱与生化参数之间的关系,并研究了利用干血清红外光谱同时量化九种主要血清成分的可行性。首先,对光谱数据与生化参数进行相关性分析,并根据相关性结果确定葡萄糖、蛋白质和脂质的相关光谱带。随后,利用区间随机蛙(IRF)算法选择相关谱带的最佳特征波数,提取信息量最大的光谱变量,构建偏最小二乘法(PLS)定量模型。该方法成功实现了对血清中葡萄糖、总蛋白、白蛋白、载脂蛋白A1、载脂蛋白B、总胆固醇、甘油三酯、低密度脂蛋白胆固醇和高密度脂蛋白胆固醇等九种主要成分的快速准确定量。实验结果表明,测试集的相关系数(Rp)范围为 0.8892-0.9941。其中,总胆固醇的定量相关系数最高,测试集中的均方根误差(RMSEP)为 7.2425 mg/dL,而葡萄糖的定量相关系数最低,相关均方根误差为 2.3683 mg/dL。本研究开发的相关分析(CA)-IRF-PLS 方法优于传统的 PLS 方法、直接使用连续投影算法(SPA)-PLS 定量方法和其他已报道的定量技术,为实时测定血清中的临床参数提供了一种新方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
8.40
自引率
11.40%
发文量
1364
审稿时长
40 days
期刊介绍:
Spectrochimica Acta, Part A: Molecular and Biomolecular Spectroscopy (SAA) is an interdisciplinary journal which spans from basic to applied aspects of optical spectroscopy in chemistry, medicine, biology, and materials science.
The journal publishes original scientific papers that feature high-quality spectroscopic data and analysis. From the broad range of optical spectroscopies, the emphasis is on electronic, vibrational or rotational spectra of molecules, rather than on spectroscopy based on magnetic moments.
Criteria for publication in SAA are novelty, uniqueness, and outstanding quality. Routine applications of spectroscopic techniques and computational methods are not appropriate.
Topics of particular interest of Spectrochimica Acta Part A include, but are not limited to:
Spectroscopy and dynamics of bioanalytical, biomedical, environmental, and atmospheric sciences,
Novel experimental techniques or instrumentation for molecular spectroscopy,
Novel theoretical and computational methods,
Novel applications in photochemistry and photobiology,
Novel interpretational approaches as well as advances in data analysis based on electronic or vibrational spectroscopy.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


