Tuning the performance of a TphR-based terephthalate biosensor with a design of experiments approach
IF 4.1
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
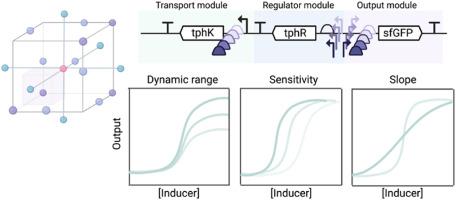
Transcription factor-based biosensors are genetic tools that aim to predictability link the presence of a specific input stimuli to a tailored gene expression output. The performance characteristics of a biosensor fundamentally determines its potential applications. However, current methods to engineer and optimise tailored biosensor responses are highly nonintuitive, and struggle to investigate multidimensional sequence/design space efficiently. In this study we employ a design of experiments (DoE) approach to build a framework for efficiently engineering activator-based biosensors with tailored performances, and we apply the framework for the development of biosensors for the polyethylene terephthalate (PET) plastic degradation monomer terephthalate (TPA). We simultaneously engineer the core promoter and operator regions of the responsive promoter, and by employing a dual refactoring approach, we were able to explore an enhanced biosensor design space and assign their causative performance effects. The approach employed here serves as a foundational framework for engineering transcriptional biosensors and enabled development of tailored biosensors with enhanced dynamic range and diverse signal output, sensitivity, and steepness. We further demonstrate its applicability on the development of tailored biosensors for primary screening of PET hydrolases and enzyme condition screening, demonstrating the potential of statistical modelling in optimising biosensors for tailored industrial and environmental applications.
用实验设计方法调整基于 TphR 的对苯二甲酸盐生物传感器的性能
基于转录因子的生物传感器是一种基因工具,旨在预测特定输入刺激与定制基因表达输出之间的联系。生物传感器的性能特征从根本上决定了其潜在的应用领域。然而,目前设计和优化定制生物传感器响应的方法非常不直观,难以有效地研究多维序列/设计空间。在本研究中,我们采用实验设计(DoE)方法建立了一个框架,用于有效地设计具有定制性能的基于激活剂的生物传感器,并将该框架用于开发聚对苯二甲酸乙二醇酯(PET)塑料降解单体对苯二甲酸乙二醇酯(TPA)的生物传感器。我们同时设计了响应型启动子的核心启动子和操作子区域,并通过采用双重重构方法,探索了增强型生物传感器的设计空间,并分配了它们的因果性能效应。本文采用的方法可作为转录生物传感器工程设计的基础框架,并能开发出动态范围更广、信号输出、灵敏度和陡度更多样化的定制生物传感器。我们进一步证明了该方法在开发用于 PET水解酶初筛和酶条件筛选的定制生物传感器方面的适用性,展示了统计建模在优化生物传感器以实现定制工业和环境应用方面的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Metabolic Engineering Communications
Medicine-Endocrinology, Diabetes and Metabolism
CiteScore
13.30
自引率
1.90%
发文量
22
审稿时长
18 weeks
期刊介绍:
Metabolic Engineering Communications, a companion title to Metabolic Engineering (MBE), is devoted to publishing original research in the areas of metabolic engineering, synthetic biology, computational biology and systems biology for problems related to metabolism and the engineering of metabolism for the production of fuels, chemicals, and pharmaceuticals. The journal will carry articles on the design, construction, and analysis of biological systems ranging from pathway components to biological complexes and genomes (including genomic, analytical and bioinformatics methods) in suitable host cells to allow them to produce novel compounds of industrial and medical interest. Demonstrations of regulatory designs and synthetic circuits that alter the performance of biochemical pathways and cellular processes will also be presented. Metabolic Engineering Communications complements MBE by publishing articles that are either shorter than those published in the full journal, or which describe key elements of larger metabolic engineering efforts.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


