Bacterial Biosynthesis of Nitrile-Containing Natural Products: Basis for Recognition of Diversified Substrates
IF 11.3
1区 化学
Q1 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
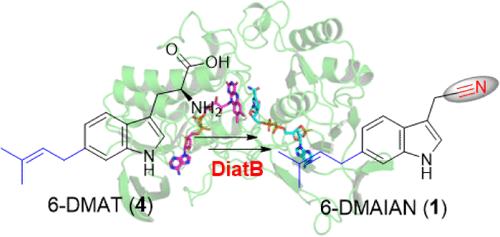
Nitrile-containing natural products, despite being a limited group of secondary metabolites, display remarkable structural and functional diversities. Aldoxime formation represents a crucial step in nitrile installation via the aldoxime-nitrile pathway although structural information regarding aldoxime formation is extremely limited. Here, we report the isolation of a nitrile compound 6-dimethylallylindole-3-acetonitrile (6-DMAIAN) and identify the aldoxime-forming enzyme gene diatB as a robust reporter for bacterial nitrile biosynthesis. We characterize the flavin-dependent monooxygenase DiatB and provide structural and mechanistic insights into the structural parameters dictating its substrate compatibilites. This enzyme initiates a nucleophilic attack on the amino group of the substrate 6-dimethylallyl-l-tryptophan (6-DMAT), resulting in formation of a transient aldoxime that precedes nitrile installation. Moreover, the DiatB recognition motif is elucidated shedding light on its substrate flexibility. We also apply bioinformatics analysis to examine the distribution and diversity of functional DiatB homologues across an array of potential nitrile-forming organisms. Given the activity of DiatB and its prevalence in secondary metabolite biosynthesis, our results provide important insight into what is, arguably, the most crucial and pivotal step in nitrile biosynthesis; these findings also suggest a promising enzymatic tool for nitrile drug design.
含腈天然产物的细菌生物合成:识别多样化底物的基础
含腈天然产物尽管是有限的次级代谢产物,但在结构和功能上却具有显著的多样性。醛肟的形成是通过醛肟-腈途径安装腈的关键步骤,但有关醛肟形成的结构信息极为有限。在此,我们报告了腈化合物 6-二甲基烯丙基吲哚-3-乙腈(6-DMAIAN)的分离情况,并确定了醛肟形成酶基因 diatB 是细菌腈生物合成的可靠报告物。我们描述了黄素依赖性单加氧酶 DiatB 的特征,并从结构和机理上深入探讨了决定其底物相容性的结构参数。这种酶对底物 6-二甲基烯丙基-l-色氨酸(6-DMAT)的氨基进行亲核攻击,形成瞬时醛肟,然后安装腈。此外,我们还阐明了 DiatB 的识别基团,揭示了其底物的灵活性。我们还应用生物信息学分析,研究了功能性 DiatB 同源物在一系列潜在腈形成生物中的分布和多样性。鉴于 DiatB 的活性及其在次生代谢物生物合成中的普遍存在,我们的研究结果为了解腈类生物合成中最关键和最重要的一步提供了重要的视角;这些发现还为腈类药物的设计提供了一种前景广阔的酶学工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Catalysis
CHEMISTRY, PHYSICAL-
CiteScore
20.80
自引率
6.20%
发文量
1253
审稿时长
1.5 months
期刊介绍:
ACS Catalysis is an esteemed journal that publishes original research in the fields of heterogeneous catalysis, molecular catalysis, and biocatalysis. It offers broad coverage across diverse areas such as life sciences, organometallics and synthesis, photochemistry and electrochemistry, drug discovery and synthesis, materials science, environmental protection, polymer discovery and synthesis, and energy and fuels.
The scope of the journal is to showcase innovative work in various aspects of catalysis. This includes new reactions and novel synthetic approaches utilizing known catalysts, the discovery or modification of new catalysts, elucidation of catalytic mechanisms through cutting-edge investigations, practical enhancements of existing processes, as well as conceptual advances in the field. Contributions to ACS Catalysis can encompass both experimental and theoretical research focused on catalytic molecules, macromolecules, and materials that exhibit catalytic turnover.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


