In vivo assembly of complete eukaryotic nucleosomes and (H3-H4)-only non-canonical nucleosomal particles in the model bacterium Escherichia coli
IF 5.2
1区 生物学
Q1 BIOLOGY
引用次数: 0
Abstract
As a fundamental unit for packaging genomic DNA into chromatin, the eukaryotic nucleosome core comprises a canonical octamer with two copies for each histone, H2A, H2B, H3, and H4, wrapped around with 147 base pairs of DNA. While H3 and H4 share structure-fold with archaeal histone-like proteins, the eukaryotic nucleosome core and the complete nucleosome (the core plus H1 histone) are unique to eukaryotes. To explore whether the eukaryotic nucleosome can assemble in prokaryotes and to reconstruct the possible route for its emergence in eukaryogenesis, we developed an in vivo system for assembly of nucleosomes in the model bacterium, Escherichia coli, and successfully reconstituted the core nucleosome, the complete nucleosome, and unexpectedly the non-canonical (H3-H4)4 octasome. The core and complete nucleosomes assembled in E. coli exhibited footprints typical of eukaryotic hosts after in situ micrococcal nuclease digestion. Additionally, they caused condensation of E. coli nucleoid. We also demonstrated the stable formation of non-canonical (H3-H4)2 tetrasome and (H3-H4)4 octasomes in vivo, which are suggested to be ‘fossil complex’ that marks the intermediate in the progressive development of eukaryotic nucleosome. The study presents a unique platform in a bacterium for in vivo assembly and studying the properties of non-canonical variants of nucleosome. Development of an in vivo system for assembly of eukaryotic nucleosomes in Escherichia coli, including the non-canonical (H3-H4)4 octasome, suggests the H3-H4 tetrasome and octasomes are likely intermediate complexes in the evolution of the eukaryotic nucleosome.
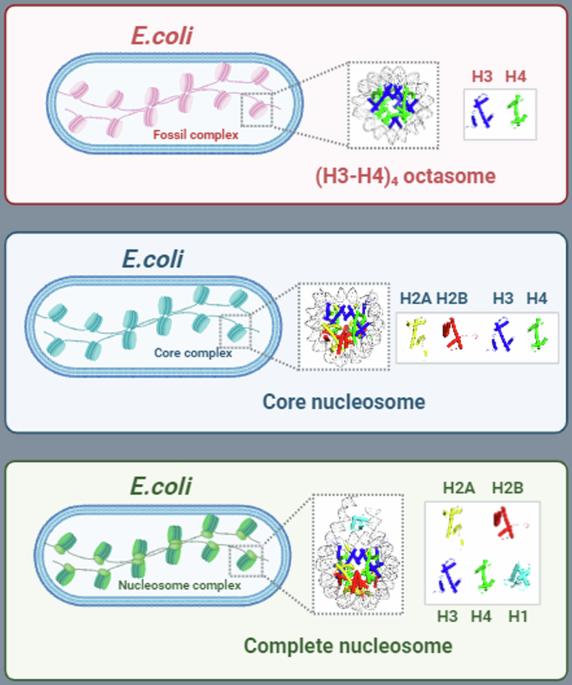
在模式菌大肠杆菌体内组装完整的真核生物核小体和仅含(H3-H4)的非规范核小体颗粒。
作为将基因组 DNA 包装成染色质的基本单位,真核生物核小体核心由一个典型的八聚体组成,每个组蛋白(H2A、H2B、H3 和 H4)都有两个拷贝,外面包着 147 个碱基对的 DNA。虽然 H3 和 H4 与古生组蛋白类似蛋白具有相同的结构折叠,但真核生物核小体核心和完整的核小体(核心加 H1 组蛋白)却是真核生物所独有的。为了探索真核生物核小体能否在原核生物中组装,并重建其在真核发生过程中出现的可能途径,我们在模式细菌大肠杆菌中开发了一个体内核小体组装系统,并成功地重组了核心核小体、完整核小体以及非经典的(H3-H4)4八聚体。在大肠杆菌中组装的核心核小体和完整核小体在原位微球菌核酸酶消化后显示出真核宿主的典型足迹。此外,它们还引起了大肠杆菌核小体的凝集。我们还证明了非经典(H3-H4)2 四体和(H3-H4)4 八体在体内的稳定形成,它们被认为是 "化石复合体",标志着真核生物核小体逐步发展的中间过程。这项研究提供了一个独特的细菌平台,用于在体内组装和研究核小体非典型变体的特性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


