Accounting for heterogeneity due to environmental sources in meta-analysis of genome-wide association studies
IF 5.2
1区 生物学
Q1 BIOLOGY
引用次数: 0
Abstract
Meta-analysis of genome-wide association studies (GWAS) across diverse populations offers power gains to identify loci associated with complex traits and diseases. Often heterogeneity in effect sizes across populations will be correlated with genetic ancestry and environmental exposures (e.g. lifestyle factors). We present an environment-adjusted meta-regression model (env-MR-MEGA) to detect genetic associations by adjusting for and quantifying environmental and ancestral heterogeneity between populations. In simulations, env-MR-MEGA has similar or greater association power than MR-MEGA, with notable gains when the environmental factor has a greater correlation with the trait than ancestry. In our analysis of low-density lipoprotein cholesterol in ~19,000 individuals across twelve sex-stratified GWAS from Africa, adjusting for sex, BMI, and urban status, we identify additional heterogeneity beyond ancestral effects for seven variants. Env-MR-MEGA provides an approach to account for environmental effects using summary-level data, making it a useful tool for meta-analyses without the need to share individual-level data. Adjusting for and quantifying environmental heterogeneity in the meta-analysis of genomewide association studies of diverse populations identifies additional heterogeneity beyond ancestral effects.
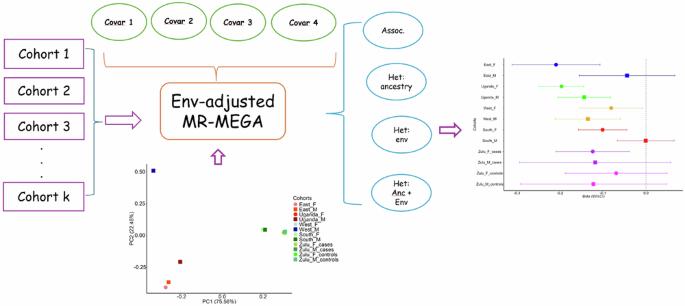
在全基因组关联研究的荟萃分析中考虑环境因素导致的异质性。
对不同人群的全基因组关联研究(GWAS)进行元分析可提高识别与复杂性状和疾病相关的基因位点的能力。不同人群间效应大小的异质性往往与遗传血统和环境暴露(如生活方式因素)相关。我们提出了一种环境调整元回归模型(env-MR-MEGA),通过调整和量化人群间的环境和祖先异质性来检测遗传关联。在模拟实验中,env-MR-MEGA 的关联能力与 MR-MEGA 相似或更强,当环境因素与性状的相关性大于祖先相关性时,env-MR-MEGA 的关联能力会显著提高。我们对非洲 12 个性别分层 GWAS 中约 19,000 人的低密度脂蛋白胆固醇进行了分析,并对性别、体重指数和城市状况进行了调整。Env-MR-MEGA提供了一种利用汇总级数据解释环境效应的方法,使其成为一种有用的荟萃分析工具,而无需共享个体级数据。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


